This is a preprint.
Cytoplasmic ribosomes hitchhike on mitochondria to dendrites
- PMID: 39314452
- PMCID: PMC11419105
- DOI: 10.1101/2024.09.13.612863
Cytoplasmic ribosomes hitchhike on mitochondria to dendrites
Abstract
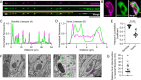
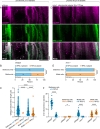
Neurons rely on local protein synthesis to rapidly modify the proteome of neurites distant from the cell body. A prerequisite for local protein synthesis is the presence of ribosomes in the neurite, but the mechanisms of ribosome transport in neurons remain poorly defined. Here, we find that ribosomes hitchhike on mitochondria for their delivery to the dendrite of a sensory neuron in C. elegans. Ribosomes co-transport with dendritic mitochondria, and their association requires the atypical Rho GTPase MIRO-1. Disrupting mitochondrial transport prevents ribosomes from reaching the dendrite, whereas ectopic re-localization of mitochondria results in a concomitant re-localization of ribosomes, demonstrating that mitochondria are required and sufficient for instructing ribosome distribution in dendrites. Endolysosomal organelles that are involved in mRNA transport and translation can associate with mitochondria and ribosomes but do not play a significant role in ribosome transport. These results reveal a mechanism for dendritic ribosome delivery, which is a critical upstream requirement for local protein synthesis.
Figures




References
-
- Baumann S.J., Grawenhoff J., Rodrigues E.C., Speroni S., Gili M., Komissarov A., and Maurer S.P.. 2022. APC couples neuronal mRNAs to multiple kinesins, EB1, and shrinking microtubule ends for bidirectional mRNA motility. Proc. Natl. Acad. Sci. 119:e2211536119. doi:10.1073/pnas.2211536119. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
