This is a preprint.
Quantitative cytoarchitectural phenotyping of deparaffinized human brain tissues
- PMID: 39314456
- PMCID: PMC11419081
- DOI: 10.1101/2024.09.10.612232
Quantitative cytoarchitectural phenotyping of deparaffinized human brain tissues
Abstract
Advanced 3D imaging techniques and image segmentation and classification methods can profoundly transform biomedical research by offering deep insights into the cytoarchitecture of the human brain in relation to pathological conditions. Here, we propose a comprehensive pipeline for performing 3D imaging and automated quantitative cellular phenotyping on Formalin-Fixed Paraffin-Embedded (FFPE) human brain specimens, a valuable yet underutilized resource. We exploited the versatility of our method by applying it to different human specimens from both adult and pediatric, normal and abnormal brain regions. Quantitative data on neuronal volume, ellipticity, local density, and spatial clustering level were obtained from a machine learning-based analysis of the 3D cytoarchitectural organization of cells identified by different molecular markers in two subjects with malformations of cortical development (MCD). This approach will grant access to a wide range of physiological and pathological paraffin-embedded clinical specimens, allowing for volumetric imaging and quantitative analysis of human brain samples at cellular resolution. Possible genotype-phenotype correlations can be unveiled, providing new insights into the pathogenesis of various brain diseases and enlarging treatment opportunities.
Figures

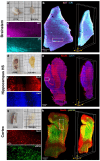
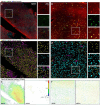

Similar articles
-
DNA methylation-based classification of malformations of cortical development in the human brain.Acta Neuropathol. 2022 Jan;143(1):93-104. doi: 10.1007/s00401-021-02386-0. Epub 2021 Nov 19. Acta Neuropathol. 2022. PMID: 34797422 Free PMC article.
-
Formalin-Fixed, Paraffin-Embedded Tissues (FFPE) as a Robust Source for the Profiling of Native and Protease-Generated Protein Amino Termini.Mol Cell Proteomics. 2016 Jun;15(6):2203-13. doi: 10.1074/mcp.O115.056515. Epub 2016 Apr 17. Mol Cell Proteomics. 2016. PMID: 27087653 Free PMC article.
-
MR microscopy of formalin fixed paraffin embedded histology specimens.Magn Reson Med. 2014 Jun;71(6):1989-94. doi: 10.1002/mrm.25225. Epub 2014 Apr 8. Magn Reson Med. 2014. PMID: 24715442
-
Applications of mass spectrometry for quantitative protein analysis in formalin-fixed paraffin-embedded tissues.Proteomics. 2014 Mar;14(4-5):441-51. doi: 10.1002/pmic.201300311. Proteomics. 2014. PMID: 24339433 Free PMC article. Review.
-
Computational solutions for spatial transcriptomics.Comput Struct Biotechnol J. 2022 Sep 1;20:4870-4884. doi: 10.1016/j.csbj.2022.08.043. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36147664 Free PMC article. Review.
References
-
- Banik G, Betts CB, Liudahl SM, Sivagnanam S, Kawashima R, Cotechini T, Larson W, Goecks J, Pai SI, Clayburgh DR, Tsujikawa T, Coussens LM (2020) High-dimensional multiplexed immunohistochemical characterization of immune contexture in human cancers. In: Methods in Enzymology. Elsevier, pp 1–20 - PMC - PubMed
-
- Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Rudy M, Eren K, Cervantes JI, Xu B, Beuttenmueller F, Wolny A, Zhang C, Koethe U, Hamprecht FA, Kreshuk A (2019) ilastik: interactive machine learning for (bio)image analysis. Nat Methods 16:1226–1232. doi: 10.1038/s41592-019-0582-9 - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
