This is a preprint.
Track-A-Worm 2.0: A Software Suite for Quantifying Properties of C. elegans Locomotion, Bending, Sleep, and Action Potentials
- PMID: 39314462
- PMCID: PMC11418985
- DOI: 10.1101/2024.09.12.612524
Track-A-Worm 2.0: A Software Suite for Quantifying Properties of C. elegans Locomotion, Bending, Sleep, and Action Potentials
Abstract
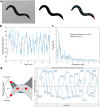
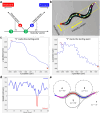
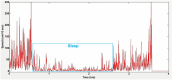
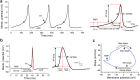
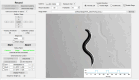
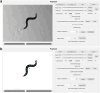
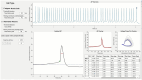
Comparative analyses of locomotor behavior and cellular electrical properties between wild-type and mutant C. elegans are crucial for exploring the gene basis of behaviors and the underlying cellular mechanisms. Although many tools have been developed by research labs and companies, their application is often hindered by implementation difficulties or lack of features specifically suited for C. elegans. Track-A-Worm 2.0 addresses these challenges with three key components: WormTracker, SleepTracker, and Action Potential (AP) Analyzer. WormTracker accurately quantifies a comprehensive set of locomotor and body bending metrics, reliably distinguish between the ventral and dorsal sides, continuously tracks the animal using a motorized stage, and seamlessly integrates external devices, such as a light source for optogenetic stimulation. SleepTracker detects and quantifies sleep-like behavior in freely moving animals. AP Analyzer assesses the resting membrane potential, afterhyperpolarization level, and various AP properties, including threshold, amplitude, mid-peak width, rise and decay times, and maximum and minimum slopes. Importantly, it addresses the challenge of AP threshold quantification posed by the absence of a pre-upstroke inflection point. Track-A-Worm 2.0 is potentially a valuable tool for many C. elegans research labs due to its powerful functionality and ease of implementation.
Figures





Similar articles
-
Track-a-worm, an open-source system for quantitative assessment of C. elegans locomotory and bending behavior.PLoS One. 2013 Jul 26;8(7):e69653. doi: 10.1371/journal.pone.0069653. Print 2013. PLoS One. 2013. PMID: 23922769 Free PMC article.
-
Comparative Analysis of Experimental Methods to Quantify Animal Activity in Caenorhabditis elegans Models of Mitochondrial Disease.J Vis Exp. 2021 Apr 4;(170):10.3791/62244. doi: 10.3791/62244. J Vis Exp. 2021. PMID: 33871460 Free PMC article.
-
Automatically tracking feeding behavior in populations of foraging C. elegans.Elife. 2022 Sep 9;11:e77252. doi: 10.7554/eLife.77252. Elife. 2022. PMID: 36083280 Free PMC article.
-
Call it Worm Sleep.Trends Neurosci. 2016 Feb;39(2):54-62. doi: 10.1016/j.tins.2015.12.005. Epub 2015 Dec 30. Trends Neurosci. 2016. PMID: 26747654 Free PMC article. Review.
-
Microfluidics as a tool for C. elegans research.WormBook. 2013 Sep 24:1-19. doi: 10.1895/wormbook.1.162.1. WormBook. 2013. PMID: 24065448 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
