Development of a RPA-CRISPR/Cas12a based rapid visual detection assay for Porcine Parvovirus 7
- PMID: 39315085
- PMCID: PMC11417039
- DOI: 10.3389/fvets.2024.1440769
Development of a RPA-CRISPR/Cas12a based rapid visual detection assay for Porcine Parvovirus 7
Abstract
Introduction: Porcine Parvovirus (PPV) is a significant pathogen in the pig industry, with eight genotypes, including PPV7, identified since its emergence in 2016. Co-infections with viruses such as Porcine Circovirus 2 (PCV2) and Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) pose serious risks to swine health. Thus, there is an urgent need for rapid, sensitive, and specific detection methods suitable for use in field settings or laboratories with limited resources.
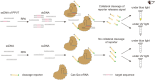
Methods: We developed a CRISPR/Cas12a-based assay combined with recombinase polymerase amplification (RPA) for the rapid detection of PPV7. Specific RPA primers and five CRISPR RNAs (crRNAs) were designed to target a highly conserved region within the NS1 gene of PPV7. Optimization of crRNA and single-stranded DNA (ssDNA) concentrations was performed to enhance the assay's performance.
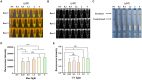
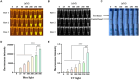
Results: CrRNA optimization identified crRNA-05 as the optimal candidate for Cas12a-based detection of PPV7, as all synthesized crRNAs demonstrated similar performance. The optimal crRNA concentration was determined to be 200 nM, yielding consistent results across tested concentrations. For ssDNA optimization, the strongest fluorescence signal was achieved with 500 nM of the FAM-BHQ ssDNA receptor. The assay showed a minimal detection limit of 100copies/μl for PPV7, confirmed through fluorescence and lateral flow detection methods. Specificity testing indicated that only PPV7 DNA samples returned positive results, confirming the assay's accuracy. In tests of 50 lung tissue samples from diseased pigs, the RPA-Cas12a assay identified 29 positive samples (58%), surpassing the 22 positive samples (44%) detected by conventional PCR. This highlights the RPA-Cas12a method's enhanced detection capability and its potential utility in clinical surveillance and management of PPV7 in swine populations.
Discussion: The RPA-Cas12a assay effectively detects PPV7 in clinical samples, enhancing disease surveillance and control in pigs. Its adaptability to resource-limited settings significantly improves PPV7 management and prevention strategies, thereby supporting the overall health and development of the pig industry.
Keywords: CRISPR/Cas12a; PCR; Porcine Parvovirus 7; detection; visual.
Copyright © 2024 Wen, She, Dang, Liao, Li, Zhang, Song, Li and Zhai.
Conflict of interest statement
AL was employed by Guangzhou Yitun Pig Industry Co. Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
KP177R-based visual assay integrating RPA and CRISPR/Cas12a for the detection of African swine fever virus.Front Immunol. 2024 Apr 9;15:1358960. doi: 10.3389/fimmu.2024.1358960. eCollection 2024. Front Immunol. 2024. PMID: 38655256 Free PMC article.
-
Rapid and Visual Detection of Porcine Parvovirus Using an ERA-CRISPR/Cas12a System Combined With Lateral Flow Dipstick Assay.Front Cell Infect Microbiol. 2022 May 11;12:879887. doi: 10.3389/fcimb.2022.879887. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35646725 Free PMC article.
-
Prevalence of porcine parvovirus 1 through 7 (PPV1-PPV7) and co-factor association with PCV2 and PRRSV in Korea.BMC Vet Res. 2022 Apr 9;18(1):133. doi: 10.1186/s12917-022-03236-1. BMC Vet Res. 2022. PMID: 35395853 Free PMC article.
-
Ov-RPA-CRISPR/Cas12a assay for the detection of Opisthorchis viverrini infection in field-collected human feces.Parasit Vectors. 2024 Feb 21;17(1):80. doi: 10.1186/s13071-024-06134-7. Parasit Vectors. 2024. PMID: 38383404 Free PMC article.
-
Sensitive and visual detection of SARS-CoV-2 using RPA-Cas12a one-step assay with ssDNA-modified crRNA.Anal Chim Acta. 2024 Jun 22;1309:342693. doi: 10.1016/j.aca.2024.342693. Epub 2024 May 5. Anal Chim Acta. 2024. PMID: 38772660
References
LinkOut - more resources
Full Text Sources

