Etiological stratification and prognostic assessment of haemophagocytic lymphohistiocytosis by machine learning on onco-mNGS data and clinical data
- PMID: 39315095
- PMCID: PMC11416948
- DOI: 10.3389/fimmu.2024.1390298
Etiological stratification and prognostic assessment of haemophagocytic lymphohistiocytosis by machine learning on onco-mNGS data and clinical data
Abstract
Introduction: Hemophagocytic lymphohistiocytosis (HLH) is a rare, complicated and life threatening hyperinflammatory syndrome that maybe triggered by various infectious agents, malignancies and rheumatologic disorders. Early diagnosis and identification of the cause is essential to initiate appropriate treatment and improve the quality of life and survival of patients. The recently developed Onco-mNGS technology can be successfully used for simultaneous detection of infections and tumors.
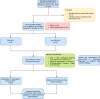
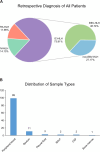
Methods: In the present study, 92 patients with clinically confirmed HLH were etiologically subtyped for infection, tumor and autoimmunity based on CNV and microbial data generated by Onco-mNGS technology, and a predictive model was developed and validated for the differential diagnosis of the underlying disease leading to secondary HLH. Furthermore, the treatment outcomes of patients with HLH triggered by EBV infection and non-EBV infection were evaluated, respectively.
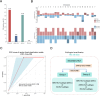
Results: The current study demonstrated that the novel Onco-mNGS can identify the infection and malignancy- related triggers among patients with secondary HLH. A random forest classification model based on CNV profile, infectious pathogen spectrum and blood microbial community was developed to better identify the different HLH subtypes and determine the underlying triggers. The prognosis for treatment of HLH patients is not only associated with CNV, but also with the presence of pathogens and non- pathogens in peripheral blood. Higher CNV burden along with frequent deletions on chromosome 19, higher pathogen burden and lower non-pathogenic microbes were prognosis factors that significantly related with unfavorable treatment outcomes.
Discussion: Our study provided comprehensive knowledge in the triggers and prognostic predictors of patients with secondary HLH, which may help early diagnosis and appropriate targeted therapy, thus improving the survival and prognosis of the patients.
Keywords: etiological stratification; hemophagocytic lymphohistiocytosis; machine learning; onco-mNGS; prognostic assessment.
Copyright © 2024 Wu, Cao, Wang, Kong, Hu, Shi, Dou, Song, Chen, Zhou, Liu, Ren and Wang.
Conflict of interest statement
Author XC is employed by MatriDx Biotechnology Co., Ltd. Author RR is employed by EBV-Care Biotechnology Co., Ltd., Micro-Health Biotechnology Co., Ltd. and Foshan Branch, Institute of Biophysics, Chinese Academy of Sciences. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources

