This is a preprint.
Cytochrome c oxidase dependent respiration is essential for T cell activation, proliferation and memory formation
- PMID: 39315267
- PMCID: PMC11419267
- DOI: 10.21203/rs.3.rs-4875322/v2
Cytochrome c oxidase dependent respiration is essential for T cell activation, proliferation and memory formation
Abstract
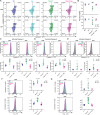
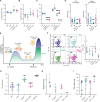
T cell activation, proliferation, and differentiation are fundamentally driven by shifts in cellular metabolism, with mitochondria playing a central role. Cytochrome c oxidase (COX, complex IV) is a key player in this process, as its activity is crucial for apoptosis, mtDNA maintenance, mitochondrial transcription, and mitochondrial respiration (MR), all of which influence T cell fate and function. Despite its known roles, the specific functions of COX required for T cell activity in vivo remain unclear. To isolate the role of MR in T cell function, we reintroduced this capability in COX-deficient T cells using an alternative oxidase (AOX) from Ciona intestinalis. Our findings demonstrate that MR is vital for maintaining metabolic balance during T cell activation by alleviating electron pressure from metabolic reprogramming and preserving redox homeostasis. We further showed that AOX mitigates apoptosis, prevents metabolic disruptions in glycolysis and the tricarboxylic acid cycle, and improves mtDNA maintenance and transcription, indicating that these disturbances are secondary to impaired MR in the absence of COX. Most importantly, the introduction of AOX restored robust effector and memory T cell generation and function in COX-deficient cells. These results highlight the essential role of COX-dependent MR in ensuring cellular health and underscore its pivotal role in T cell proliferation and differentiation.
Conflict of interest statement
Competing interests: MSZ is a co-founder of a start-up company founded to develop therapeutics based on AOX. Additional Declarations: There is NO Competing Interest.
Figures




References
-
- Miranda M., Bonekamp N.A. & Kühl I. Starting the engine of the powerhouse: mitochondrial transcription and beyond. Biological Chemistry 403, 779–805 (2022). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

