De novo and inherited micro-CNV at 16p13.11 in 21 Chinese patients with defective cardiac left-right patterning
- PMID: 39315310
- PMCID: PMC11416941
- DOI: 10.3389/fgene.2024.1458953
De novo and inherited micro-CNV at 16p13.11 in 21 Chinese patients with defective cardiac left-right patterning
Abstract
Objective: Copy number changes at Chromosomal 16p13.11 have been implicated in a variety of human diseases including congenital cardiac abnormalities. The clinical correlation of copy number variants (CNVs) in this region with developmental abnormalities remains controversial as most of the patients inherit the duplication from an unaffected parent.
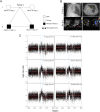
Methods: We performed CNV analysis on 164 patients with defective left-right (LR) patterning based on whole genome-exome sequencing (WG-ES) followed by multiplex ligation-dependent probe amplification (MLPA) validation. Most cases were accompanied with complex congenital heart disease (CHD).
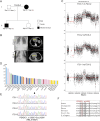
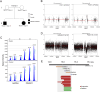
Results: CNVs at 16p13.11 were identified in a total of 21 cases, accounting for 12.80% (21/164) evaluated cases. We observed a marked overrepresentation of chromosome 16p13.11 duplications in cases when compared with healthy controls according to literature reports (15/164, 9.14% versus 0.09% in controls). Notably, in two independent family trios, de novo 16p13.11 micro-duplications were identified in two patients with laterality defects and CHD. Moreover, 16p13.11 micro-duplication was segregated with the disease in a family trio containing 2 affected individuals. Notably, five coding genes, NOMO1, PKD1P3, NPIPA1, PDXDC1, and NTAN1, were potentially affected by micro-CNV at 16p13.11 in these patients.
Conclusion: Our study provides new family-trio based evidences to support 16p13.11 micro-duplications predispose individuals to defective cardiac left-right patterning and laterality disorder.
Keywords: 16p13.11; ciliopathy; congenital heart disease; copy number variation; deletion; duplication; laterality disorder; left-right patterning.
Copyright © 2024 Yu, Chen, Chen, Shen, Wu, Zhang and Zhou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Allach El Khattabi L., Heide S., Caberg J. H., Andrieux J., Doco Fenzy M., Vincent-Delorme C., et al. (2018). 16p13.11 microduplication in 45 new patients: refined clinical significance and genotype-phenotype correlations. J. Med. Genet. 57, 301–307. pii: jmedgenet-2018-105389. 10.1136/jmedgenet-2018-105389 - DOI - PubMed
-
- Bartoloni L., Blouin J. L., Pan Y., Gehrig C., Maiti A. K., Scamuffa N., et al. (2002). Mutations in the DNAH11 (axonemal heavy chain dynein type 11) gene cause one form of situs inversus totalis and most likely primary ciliary dyskinesia. Proc. Natl. Acad. Sci. U. S. A. 99 (16), 10282–10286. 10.1073/pnas.152337699 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

