Distinct molecular features of FLNC mutations, associated with different clinical phenotypes
- PMID: 39315490
- PMCID: PMC11904857
- DOI: 10.1002/cm.21922
Distinct molecular features of FLNC mutations, associated with different clinical phenotypes
Abstract
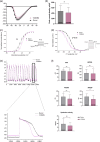
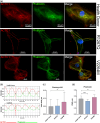
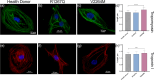
Filamin С is a key an actin-binding protein of muscle cells playing a critical role in maintaining structural integrity and sarcomere organization. FLNC mutations contribute to various types of cardiomyopathies and myopathies through potentially different molecular mechanisms. Here, we described the impact of two clinically distinct FLNC variants (R1267Q associated with arrhythmogenic cardiomyopathy and V2264M associated with restrictive cardiomyopathy) on calcium homeostasis, electrophysiology, and gene expression profile of iPSC-derived patient-specific cardiomyocytes. We demonstrated that R1267Q FLNC variant leads to greater disturbances in calcium dynamics, Nav1.5 kinetics and action potentials compared to V2264M variant. These functional characteristics were accompanied by transcriptome changes in genes linked to action potential and sodium transport as well as structural cardiomyocyte genes. We suggest distinct molecular effects of two FLNC variants linked to different types of cardiomyopathies in terms of myofilament structure, electrophysiology, ion channel function and intracellular calcium homeostasis providing the molecular the bases for their different clinical phenotypes.
Keywords: cardiomyocyte; cardiomyopathy; electrophysiology; filamin; iPSC; transcriptome.
© 2024 The Author(s). Cytoskeleton published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

