Real-world assessment of comprehensive genome profiling impact on clinical outcomes: A single-institution study in Japan
- PMID: 39315676
- PMCID: PMC11420830
- DOI: 10.1002/cam4.70249
Real-world assessment of comprehensive genome profiling impact on clinical outcomes: A single-institution study in Japan
Abstract
Introduction: Comprehensive genome profiling (CGP) has revolutionized healthcare by offering personalized medicine opportunities. However, its real-world utility and impact remain incompletely understood. This study examined the extent to which CGP leads to genomically matched therapy and its effectiveness.
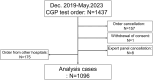
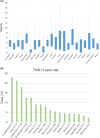
Methods: We analyzed data from advanced solid tumor patients who underwent CGP panel between December 2019 and May 2023 at the Osaka International Cancer Institute. Patient demographics, specimen details, and expert panel assessments were collected. Turnaround time (TAT) and genomically matched therapy outcomes were analyzed. Gene alterations and their co-occurrence patterns were also assessed.
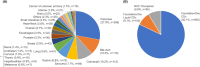
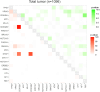
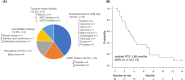
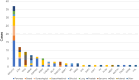
Results: Among 1437 patients, 1096 results were available for analysis. The median TAT was 63 [28-182] days. There were 667 (60.9%) cases wherein recommended clinical trials were presented and there were 12 (1.1%) cases that could be enrolled in the trial and 25 (2.3%) cases that could lead to therapies under insurance reimbursement. The median progression free survival of the trial treatment was 1.58 months (95% CI: 0.66-4.37) in clinical trials and 3.66 months (95% CI: 2.14-7.13) in treatment under insurance. Pathologic germline variants were confirmed in 15 patients (1.3%). Co-alteration of CDKN2A, CDKN2B, and MTAP was significantly observed in overall population.
Conclusion: The effectiveness of the genomically matched therapy based on the CGP panel was unsatisfactory. Expansion of clinical trials and utilization of remote clinical trials are required to ensure that the results of the CGP panel can be fully returned to patients.
Keywords: clinical trial; comprehensive genomic profiling; genomically matched therapy; real world data; solid tumor.
© 2024 The Author(s). Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
Dr. Kunimasa reports honoraria for lecture from Chugai Pharma and Dr. Nishimura reports honoraria for lectures from Astellas, AstraZeneca, MSD, Janssen, Merck Biopharma and Bayer. The other authors have no conflict of interest.
Figures

References
-
- Wahida A, Buschhorn L, Fröhling S, et al. The coming decade in precision oncology: six riddles. Nat Rev Cancer. 2023;23:43‐54. - PubMed
-
- Fulton‐Ward T, Middleton G. The impact of genomic context on outcomes of solid cancer patients treated with genotype‐matched targeted therapies: a comprehensive review. Ann Oncol. 2023;34:1113‐1130. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

