Growth defect of domain III glycoprotein B mutants of human cytomegalovirus reverted by compensatory mutations co-localizing in post-fusion conformation
- PMID: 39315800
- PMCID: PMC11481916
- DOI: 10.1128/mbio.01812-24
Growth defect of domain III glycoprotein B mutants of human cytomegalovirus reverted by compensatory mutations co-localizing in post-fusion conformation
Abstract
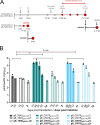
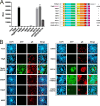
Cell entry is a crucial step for a virus to infect a host cell. Human cytomegalovirus utilizes glycoprotein B (gB) to fuse the viral and host cell membranes upon receptor binding of gH/gL-containing complexes. Fusion is mediated by major conformational changes of gB from a metastable pre-fusion to a stable post-fusion state whereby the central trimeric coiled-coils, formed by domain (Dom)III α helices, remain structurally nearly unchanged. To better understand the role of the stable core, we individually introduced three potentially helix-breaking or one disulfide bond-breaking mutation in the DIII α3 to study different aspects of the viral behavior upon long-term culturing. Two of the three helix-breaking mutations, gB_Y494P and gB_I495P, were lethal for the virus in either fibroblasts or epithelial cells. The third substitution, gB_G493P, on the other hand, displayed a delayed replication and spread, which was more pronounced in epithelial cells, hinting at an impaired fusion. Interestingly, the disulfide bond-breaker mutation, gB_C507S, performed strikingly differently in the two cell types - lethal in epithelial cells and an atypical phenotype in fibroblasts, respectively. Replication curve analyses paired with the infection efficiency, the spread morphology, and the cell-cell fusogenicity suggest a dysregulated fusion process, which could be reverted by second-site mutations mapping predominantly to gB DomV. Our findings underline the functional importance of a stable DomIII core for a well-regulated DomV rearrangement during fusion.IMPORTANCEHuman cytomegalovirus (HCMV) can establish a lifelong infection. In most people, the infection follows an asymptomatic course; however, it is a major cause of morbidity and mortality in immunocompromised patients or neonates. HCMV has a very broad cell tropism, ranging from fibroblasts to epi- and endothelial cells. The virus uses different entry pathways utilizing the core fusion machinery consisting of glycoprotein complexes gH/gL and glycoprotein B (gB). The fusion protein gB undergoes fundamental rearrangements from a metastable pre-fusion to a stable post-fusion conformation. Here, we characterized the viral behavior after the introduction of four single-point mutations in the gB central core. These led to various cell type-specific atypical phenotypes and the emergence of compensatory mutations, demonstrating an important interaction between domains III and V. We provide a new basis for the development of a structurally and functionally altered gB, which can further serve as a tool for drug and vaccine development.
Keywords: entry and spread; glycoprotein B; glycoprotein O; human cytomegalovirus; membrane fusion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

