High-throughput sequencing-based neutralization assay reveals how repeated vaccinations impact titers to recent human H1N1 influenza strains
- PMID: 39315814
- PMCID: PMC11494878
- DOI: 10.1128/jvi.00689-24
High-throughput sequencing-based neutralization assay reveals how repeated vaccinations impact titers to recent human H1N1 influenza strains
Abstract
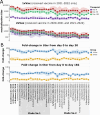
The high genetic diversity of influenza viruses means that traditional serological assays have too low throughput to measure serum antibody neutralization titers against all relevant strains. To overcome this challenge, we developed a sequencing-based neutralization assay that simultaneously measures titers against many viral strains using small serum volumes using a workflow similar to traditional neutralization assays. The key innovation is to incorporate unique nucleotide barcodes into the hemagglutinin (HA) genomic segment, and then pool viruses with numerous different barcoded HA variants and quantify the infectivity of all of them simultaneously using next-generation sequencing. With this approach, a single researcher performed the equivalent of 2,880 traditional neutralization assays (80 serum samples against 36 viral strains) in approximately 1 month. We applied the sequencing-based assay to quantify the impact of influenza vaccination on neutralization titers against recent human H1N1 strains for individuals who had or had not also received a vaccine in the previous year. We found that the viral strain specificities of the neutralizing antibodies elicited by vaccination vary among individuals and that vaccination induced a smaller increase in titers for individuals who had also received a vaccine the previous year-although the titers 6 months after vaccination were similar in individuals with and without the previous-year vaccination. We also identified a subset of individuals with low titers to a subclade of recent H1N1 even after vaccination. We provide an experimental protocol (dx.doi.org/10.17504/protocols.io.kqdg3xdmpg25/v1) and computational pipeline (https://github.com/jbloomlab/seqneut-pipeline) for the sequencing-based neutralization assays to facilitate the use of this method by others.
Importance: We describe a new approach that can rapidly measure how the antibodies in human serum inhibit infection by many different influenza strains. This new approach is useful for understanding how viral evolution affects antibody immunity. We apply the approach to study the effect of repeated influenza vaccination.
Keywords: influenza; influenza vaccines; neutralization assay; next-generation sequencing.
Conflict of interest statement
J.D.B. is on the scientific advisory boards of Apriori Bio, Invivyd, and the Vaccine Company. J.D.B. and A.N.L. receive royalty payments as inventors on Fred Hutch licensed patents related to incorporating barcodes into the influenza genome and viral deep mutational scanning. B.J.C. has consulted for AstraZeneca, Fosun Pharma, GlaxoSmithKline, Haleon, Moderna, Novavax, Pfizer, Roche, and Sanofi Pasteur. S.C. has consulted for CSL Seqirus.
Figures





Update of
-
High-throughput sequencing-based neutralization assay reveals how repeated vaccinations impact titers to recent human H1N1 influenza strains.bioRxiv [Preprint]. 2024 Mar 9:2024.03.08.584176. doi: 10.1101/2024.03.08.584176. bioRxiv. 2024. Update in: J Virol. 2024 Oct 22;98(10):e0068924. doi: 10.1128/jvi.00689-24. PMID: 38496577 Free PMC article. Updated. Preprint.
References
-
- Kim K, Vieira MC, Gouma S, Weirick ME, Hensley SE, Cobey S. 2024. Measures of population immunity can predict the dominant clade of influenza A (H3N2) in the 2017-2018 season and reveal age-associated differences in susceptibility and antibody-binding specificity. medRxiv:2023.10.26.23297569. doi:10.1101/2023.10.26.23297569 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

