Automated System to Capture Patient Symptoms From Multitype Japanese Clinical Texts: Retrospective Study
- PMID: 39316418
- PMCID: PMC11462096
- DOI: 10.2196/58977
Automated System to Capture Patient Symptoms From Multitype Japanese Clinical Texts: Retrospective Study
Abstract
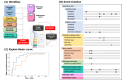
Background: Natural language processing (NLP) techniques can be used to analyze large amounts of electronic health record texts, which encompasses various types of patient information such as quality of life, effectiveness of treatments, and adverse drug event (ADE) signals. As different aspects of a patient's status are stored in different types of documents, we propose an NLP system capable of processing 6 types of documents: physician progress notes, discharge summaries, radiology reports, radioisotope reports, nursing records, and pharmacist progress notes.
Objective: This study aimed to investigate the system's performance in detecting ADEs by evaluating the results from multitype texts. The main objective is to detect adverse events accurately using an NLP system.
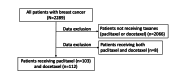
Methods: We used data written in Japanese from 2289 patients with breast cancer, including medication data, physician progress notes, discharge summaries, radiology reports, radioisotope reports, nursing records, and pharmacist progress notes. Our system performs 3 processes: named entity recognition, normalization of symptoms, and aggregation of multiple types of documents from multiple patients. Among all patients with breast cancer, 103 and 112 with peripheral neuropathy (PN) received paclitaxel or docetaxel, respectively. We evaluate the utility of using multiple types of documents by correlation coefficient and regression analysis to compare their performance with each single type of document. All evaluations of detection rates with our system are performed 30 days after drug administration.
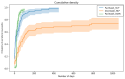
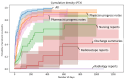
Results: Our system underestimates by 13.3 percentage points (74.0%-60.7%), as the incidence of paclitaxel-induced PN was 60.7%, compared with 74.0% in the previous research based on manual extraction. The Pearson correlation coefficient between the manual extraction and system results was 0.87 Although the pharmacist progress notes had the highest detection rate among each type of document, the rate did not match the performance using all documents. The estimated median duration of PN with paclitaxel was 92 days, whereas the previously reported median duration of PN with paclitaxel was 727 days. The number of events detected in each document was highest in the physician's progress notes, followed by the pharmacist's and nursing records.
Conclusions: Considering the inherent cost that requires constant monitoring of the patient's condition, such as the treatment of PN, our system has a significant advantage in that it can immediately estimate the treatment duration without fine-tuning a new NLP model. Leveraging multitype documents is better than using single-type documents to improve detection performance. Although the onset time estimation was relatively accurate, the duration might have been influenced by the length of the data follow-up period. The results suggest that our method using various types of data can detect more ADEs from clinical documents.
Keywords: EHR; EHRs; ML; NLP; adverse; adverse drug reaction; adverse event; cancer; detect; detecting; detection; drug; drugs; machine learning; medication; medications; named entity recognition; natural language processing; neuropathy; note; notes; oncology; peripheral neuropathy; pharmaceutic; pharmaceutical; pharmaceuticals; pharmaceutics; pharmacology; pharmacotherapy; record; records; report; reports; symptom; symptoms; text; texts; textual.
©Tomohiro Nishiyama, Ayane Yamaguchi, Peitao Han, Lis Weiji Kanashiro Pereira, Yuka Otsuki, Gabriel Herman Bernardim Andrade, Noriko Kudo, Shuntaro Yada, Shoko Wakamiya, Eiji Aramaki, Masahiro Takada, Masakazu Toi. Originally published in JMIR Medical Informatics (https://medinform.jmir.org), 24.09.2024.
Conflict of interest statement
Conflicts of Interest: MToi has received research grants from Chugai, Takeda, Pfizer, Taiho, Japan Breast Cancer Research Group Association (JBCRG), Kyoto Breast Cancer Research Network (KBCRN), Eisai, Eli-Lilly and companies, Daiichi-Sankyo, AstraZeneca, Astellas, Shimadzu, Yakult, Nippon Kayaku, AFI technology, Luxonus, Shionogi, GL Science, Sanwa Shurui; and lecture fees from Chugai, Takeda, Pfizer, Kyowa-Kirin, Taiho, Eisai, Daiichi-Sankyo, AstraZeneca, Eli Lilly and companies, MSD, Exact Science, Novartis, Shimadzu, Yakult, Nippon Kayaku、Devicore Medical Japan, Sysmex; and advisory fees from Daiichi-Sankyo, Eli Lilly and companies, BMS, Bertis, Terumo, Kansai Medical Net.
Figures


Similar articles
-
Overview of the First Natural Language Processing Challenge for Extracting Medication, Indication, and Adverse Drug Events from Electronic Health Record Notes (MADE 1.0).Drug Saf. 2019 Jan;42(1):99-111. doi: 10.1007/s40264-018-0762-z. Drug Saf. 2019. PMID: 30649735 Free PMC article. Review.
-
Leveraging Natural Language Processing and Machine Learning Methods for Adverse Drug Event Detection in Electronic Health/Medical Records: A Scoping Review.Drug Saf. 2025 Apr;48(4):321-337. doi: 10.1007/s40264-024-01505-6. Epub 2025 Jan 9. Drug Saf. 2025. PMID: 39786481 Free PMC article.
-
Detecting Adverse Drug Events in Clinical Notes Using Large Language Models.Stud Health Technol Inform. 2025 May 15;327:892-893. doi: 10.3233/SHTI250495. Stud Health Technol Inform. 2025. PMID: 40380603
-
A Deep Learning-Enabled Workflow to Estimate Real-World Progression-Free Survival in Patients With Metastatic Breast Cancer: Study Using Deidentified Electronic Health Records.JMIR Cancer. 2025 May 15;11:e64697. doi: 10.2196/64697. JMIR Cancer. 2025. PMID: 40372953 Free PMC article.
-
Extraction of Information Related to Drug Safety Surveillance From Electronic Health Record Notes: Joint Modeling of Entities and Relations Using Knowledge-Aware Neural Attentive Models.JMIR Med Inform. 2020 Jul 10;8(7):e18417. doi: 10.2196/18417. JMIR Med Inform. 2020. PMID: 32459650 Free PMC article.
Cited by
-
Large Language Models for Adverse Drug Events: A Clinical Perspective.J Clin Med. 2025 Aug 4;14(15):5490. doi: 10.3390/jcm14155490. J Clin Med. 2025. PMID: 40807108 Free PMC article. Review.
-
Identifying Adverse Events in Outpatients With Prostate Cancer Using Pharmaceutical Care Records in Community Pharmacies: Application of Named Entity Recognition.JMIR Cancer. 2025 Mar 11;11:e69663. doi: 10.2196/69663. JMIR Cancer. 2025. PMID: 40068144 Free PMC article.
References
-
- Devlin J, Chang MW, Lee K, Toutanova K. BERT: pre-training of deep bidirectional transformers for language understanding. Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers); June 2-7, 2019; Minneapolis, Minnesota. Association for Computational Linguistics; 2019. pp. 4171–4186. https://aclanthology.org/N19-1423/ - DOI
-
- Huang SC, Pareek A, Seyyedi S, Banerjee I, Lungren MP. Fusion of medical imaging and electronic health records using deep learning: a systematic review and implementation guidelines. NPJ Digit Med. 2020;3:136. doi: 10.1038/s41746-020-00341-z. doi: 10.1038/s41746-020-00341-z.341 - DOI - DOI - PMC - PubMed
-
- Morin O, Vallières M, Braunstein S, Ginart JB, Upadhaya T, Woodruff HC, Zwanenburg A, Chatterjee A, Villanueva-Meyer JE, Valdes G, Chen W, Hong JC, Yom SS, Solberg TD, Löck S, Seuntjens J, Park C, Lambin P. An artificial intelligence framework integrating longitudinal electronic health records with real-world data enables continuous pan-cancer prognostication. Nat Cancer. 2021;2(7):709–722. doi: 10.1038/s43018-021-00236-2. - DOI - PubMed
-
- Huang Kexin, Altosaar Jaan, Ranganath Rajesh. ClinicalBERT: Modeling Clinical Notes and Predicting Hospital Readmission. arXiv. doi: 10.48550/ARXIV.1904.05342. Preprint posted online April 10, 2019. - DOI
-
- Lee J, Yoon W, Kim S, Kim D, Kim S, So C, Kang J. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36(4):1234–1240. doi: 10.1093/bioinformatics/btz682. https://europepmc.org/abstract/MED/31501885 5566506 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

