Forebrain Eml1 depletion reveals early centrosomal dysfunction causing subcortical heterotopia
- PMID: 39316454
- PMCID: PMC11450323
- DOI: 10.1083/jcb.202310157
Forebrain Eml1 depletion reveals early centrosomal dysfunction causing subcortical heterotopia
Abstract
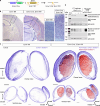
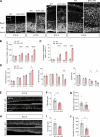
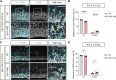
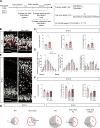
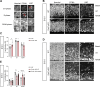
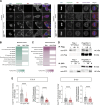
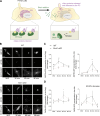
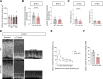
Subcortical heterotopia is a cortical malformation associated with epilepsy, intellectual disability, and an excessive number of cortical neurons in the white matter. Echinoderm microtubule-associated protein like 1 (EML1) mutations lead to subcortical heterotopia, associated with abnormal radial glia positioning in the cortical wall, prior to malformation onset. This perturbed distribution of proliferative cells is likely to be a critical event for heterotopia formation; however, the underlying mechanisms remain unexplained. This study aimed to decipher the early cellular alterations leading to abnormal radial glia. In a forebrain conditional Eml1 mutant model and human patient cells, primary cilia and centrosomes are altered. Microtubule dynamics and cell cycle kinetics are also abnormal in mouse mutant radial glia. By rescuing microtubule formation in Eml1 mutant embryonic brains, abnormal radial glia delamination and heterotopia volume were significantly reduced. Thus, our new model of subcortical heterotopia reveals the causal link between Eml1's function in microtubule regulation and cell position, both critical for correct cortical development.
© 2024 Zaidi et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures



References
-
- Bizzotto, S., Uzquiano A., Dingli F., Ershov D., Houllier A., Arras G., Richards M., Loew D., Minc N., Croquelois A., et al. . 2017. Eml1 loss impairs apical progenitor spindle length and soma shape in the developing cerebral cortex. Sci. Rep. 7:17308. 10.1038/s41598-017-15253-4 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

