GenoTriplo: A SNP genotype calling method for triploids
- PMID: 39316624
- PMCID: PMC11452025
- DOI: 10.1371/journal.pcbi.1012483
GenoTriplo: A SNP genotype calling method for triploids
Abstract
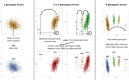
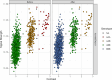
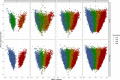
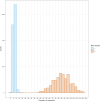
Triploidy is very useful in both aquaculture and some cultivated plants as the induced sterility helps to enhance growth and product quality, as well as acting as a barrier against the contamination of wild populations by escapees. To use genetic information from triploids for academic or breeding purposes, an efficient and robust method to genotype triploids is needed. We developed such a method for genotype calling from SNP arrays, and we implemented it in the R package named GenoTriplo. Our method requires no prior information on cluster positions and remains unaffected by shifted luminescence signals. The method relies on starting the clustering algorithm with an initial higher number of groups than expected from the ploidy level of the samples, followed by merging groups that are too close to each other to be considered as distinct genotypes. Accurate classification of SNPs is achieved through multiple thresholds of quality controls. We compared the performance of GenoTriplo with that of fitPoly, the only published method for triploid SNP genotyping with a free software access. This was assessed by comparing the genotypes generated by both methods for a dataset of 1232 triploid rainbow trout genotyped for 38,033 SNPs. The two methods were consistent for 89% of the genotypes, but for 26% of the SNPs, they exhibited a discrepancy in the number of different genotypes identified. For these SNPs, GenoTriplo had >95% concordance with fitPoly when fitPoly genotyped better. On the contrary, when GenoTriplo genotyped better, fitPoly had less than 50% concordance with GenoTriplo. GenoTriplo was more robust with less genotyping errors. It is also efficient at identifying low-frequency genotypes in the sample set. Finally, we assessed parentage assignment based on GenoTriplo genotyping and observed significant differences in mismatch rates between the best and second-best couples, indicating high confidence in the results. GenoTriplo could also be used to genotype diploids as well as individuals with higher ploidy level by adjusting a few input parameters.
Copyright: © 2024 Roche et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Nishiwaki A, Mizuguti A, Kuwabara S, Toma Y, Ishigaki G, Miyashita T, et al. Discovery of natural Miscanthus (Poaceae) triploid plants in sympatric populations of Miscanthus sacchariflorus and Miscanthus sinensis in southern Japan. American Journal of Botany. 2011. Jan 1;98(1):154–9. doi: 10.3732/ajb.1000258 - DOI - PubMed
-
- Gregory TR, Mable BK. Polyploidy in Animals. In: The Evolution of the Genome [Internet]. Elsevier; 2005. [cited 2023 Sep 12]. p. 427–517. Available from: https://linkinghub.elsevier.com/retrieve/pii/B9780123014634500103.
MeSH terms
LinkOut - more resources
Full Text Sources

