Overexpression of NUDT16L1 sustains proper function of mitochondria and leads to ferroptosis insensitivity in colorectal cancer
- PMID: 39317106
- PMCID: PMC11465047
- DOI: 10.1016/j.redox.2024.103358
Overexpression of NUDT16L1 sustains proper function of mitochondria and leads to ferroptosis insensitivity in colorectal cancer
Abstract
Cancer research is continuously exploring new avenues to improve treatments, and ferroptosis induction has emerged as a promising approach. However, the lack of comprehensive analysis of the ferroptosis sensitivity in different cancer types has limited its clinical application. Moreover, identifying the key regulator that influences the ferroptosis sensitivity during cancer progression remains a major challenge. In this study, we shed light on the role of ferroptosis in colorectal cancer and identified a novel ferroptosis repressor, NUDT16L1, that contributes to the ferroptosis insensitivity in this cancer type. Mechanistically, NUDT16L1 promotes ferroptosis insensitivity in colon cancer by enhancing the expression of key ferroptosis repressor and mitochondrial genes through direct binding to NAD-capped RNAs and the indirect action of MALAT1. Our findings also reveal that NUDT16L1 localizes to the mitochondria to maintain its proper function by preventing mitochondrial DNA leakage after treatment of ferroptosis inducer in colon cancer cells. Importantly, our orthotopic injection and Nudt16l1 transgenic mouse models of colon cancer demonstrated the critical role of NUDT16L1 in promoting tumor growth. Moreover, clinical specimens revealed that NUDT16L1 was overexpressed in colorectal cancer, indicating its potential as a therapeutic target. Finally, our study shows the therapeutic potential of a NUDT16L1 inhibitor in vitro, in vivo and ex vivo. Taken together, these findings provide new insights into the crucial role of NUDT16L1 in colorectal cancer and highlight its potential as a promising therapeutic target.
Keywords: Colon cancer; Ferroptosis insensitivity; Mitochondrial DNA leakage; Mitochondrial function; NUDT16L1; Tumor growth; metabolite-cap, MALAT1 lncRNA.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




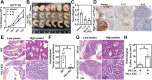
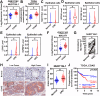
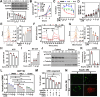
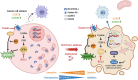
References
-
- Chen X., Kang R., Kroemer G., Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat. Rev. Clin. Oncol. 2021 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

