Specifications of the ACMG/AMP variant curation guidelines for the analysis of germline ATM sequence variants
- PMID: 39317201
- PMCID: PMC11568761
- DOI: 10.1016/j.ajhg.2024.08.022
Specifications of the ACMG/AMP variant curation guidelines for the analysis of germline ATM sequence variants
Abstract
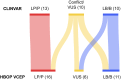
The ClinGen Hereditary Breast, Ovarian, and Pancreatic Cancer (HBOP) Variant Curation Expert Panel (VCEP) is composed of internationally recognized experts in clinical genetics, molecular biology, and variant interpretation. This VCEP made specifications for the American College of Medical Genetics and Association for Molecular Pathology (ACMG/AMP) guidelines for the ataxia telangiectasia mutated (ATM) gene according to the ClinGen protocol. These gene-specific rules for ATM were modified from the ACMG/AMP guidelines and were tested against 33 ATM variants of various types and classifications in a pilot curation phase. The pilot revealed a majority agreement between the HBOP VCEP classifications and the ClinVar-deposited classifications. Six pilot variants had conflicting interpretations in ClinVar, and re-evaluation with the VCEP's ATM-specific rules resulted in four that were classified as benign, one as likely pathogenic, and one as a variant of uncertain significance (VUS) by the VCEP, improving the certainty of interpretations in the public domain. Overall, 28 of the 33 pilot variants were not VUS, leading to an 85% classification rate. The ClinGen-approved, modified rules demonstrated value for improved interpretation of variants in ATM.
Keywords: ACMG; ATM; ClinGen; Variant Curation Expert Panel; ataxia telangiectasia; breast cancer; classification; rules specifications; variant interpretation.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.J.A. was a paid employee of Invitae. M.E.R., T.B., T.P., and C.C.Y. were paid employees of Ambry Genetics. L.Z. was a paid employee of Natera. S.H. was a paid employee of GeneDx.
Figures

Update of
-
Specifications of the ACMG/AMP variant curation guidelines for the analysis of germline ATM sequence variants.medRxiv [Preprint]. 2024 May 29:2024.05.28.24307502. doi: 10.1101/2024.05.28.24307502. medRxiv. 2024. Update in: Am J Hum Genet. 2024 Nov 7;111(11):2411-2426. doi: 10.1016/j.ajhg.2024.08.022. PMID: 38854136 Free PMC article. Updated. Preprint.
References
-
- Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/GIM.2015.30. - DOI - PMC - PubMed
-
- Plon S.E., Eccles D.M., Easton D., Foulkes W.D., Genuardi M., Greenblatt M.S., Hogervorst F.B.L., Hoogerbrugge N., Spurdle A.B., Tavtigian S.V., IARC Unclassified Genetic Variants Working Group Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 2008;29:1282–1291. doi: 10.1002/HUMU.20880. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

