Genes deregulated in giant cell arteritis by Nanostring nCounter gene expression profiling in temporal artery biopsies
- PMID: 39317454
- PMCID: PMC11423731
- DOI: 10.1136/rmdopen-2024-004600
Genes deregulated in giant cell arteritis by Nanostring nCounter gene expression profiling in temporal artery biopsies
Abstract
Objective: To identify differentially expressed genes in temporal artery biopsies (TABs) from patients with giant cell arteritis (GCA) with different histological patterns of inflammation: transmural inflammation (TMI) and inflammation limited to adventitia (ILA), compared with normal TABs from patients without GCA.
Methods: Expression of 770 immune-related genes was profiled with the NanoString nCounter PanCancer Immune Profiling Panel on formalin-fixed paraffin-embedded TABs from 42 GCA patients with TMI, 7 GCA patients with ILA and 7 non-GCA controls.
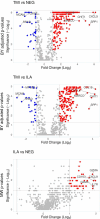
Results: Unsupervised clustering of the samples revealed two distinct groups: normal TABs and TABs with ILA in one group, 41/42 TABs with TMI in the other one. TABs with TMI showed 31 downregulated and 256 upregulated genes compared with normal TABs; they displayed 26 downregulated and 187 upregulated genes compared with TABs with ILA (>2.0 fold changes and adjusted p values <0.05). Gene expression in TABs with ILA resembled normal TABs although 38 genes exhibited >2.0 fold changes, but these changes lost statistical significance after Benjamini-Yekutieli correction. Genes encoding TNF superfamily members, immune checkpoints, chemokine and chemokine receptors, toll-like receptors, complement molecules, Fc receptors for IgG antibodies, signalling lymphocytic activation molecules, JAK3, STAT1 and STAT4 resulted upregulated in TMI.
Conclusions: TABs with TMI had a distinct transcriptome compared with normal TABs and TABs with ILA. The few genes potentially deregulated in ILA were also deregulated in TMI. Gene profiling allowed to deepen the knowledge of GCA pathogenesis.
Keywords: Autoimmune Diseases; Giant Cell Arteritis; Inflammation; Systemic vasculitis.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
