Development and application of the GenoBaits WheatSNP16K array to accelerate wheat genetic research and breeding
- PMID: 39318097
- PMCID: PMC11783889
- DOI: 10.1016/j.xplc.2024.101138
Development and application of the GenoBaits WheatSNP16K array to accelerate wheat genetic research and breeding
Abstract
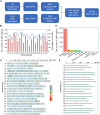
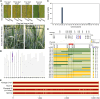
Single-nucleotide polymorphisms (SNPs) are widely used as molecular markers for constructing genetic linkage maps in wheat. Compared with available SNP-based genotyping platforms, a genotyping by target sequencing (GBTS) system with capture-in-solution (liquid chip) technology has become the favored genotyping technology because it is less demanding and more cost effective, flexible, and user-friendly. In this study, a new GenoBaits WheatSNP16K (GBW16K) GBTS array was designed using datasets generated by the wheat 660K SNP array and resequencing platforms in our previous studies. The GBW16K array contains 14 868 target SNP regions that are evenly distributed across the wheat genome, and 37 669 SNPs in these regions can be identified in a diversity panel consisting of 239 wheat accessions from around the world. Principal component and neighbor-joining analyses using the called SNPs are consistent with the pedigree information and geographic distributions or ecological environments of the accessions. For the GBW16K marker panel, the average genetic diversity among the 239 accessions is 0.270, which is sufficient for linkage map construction and preliminary mapping of targeted genes or quantitative trait loci (QTLs). A genetic linkage map, constructed using the GBW16K array-based genotyping of a recombinant inbred line population derived from a cross of the CIMMYT wheat line Yaco"S" and the Chinese landrace Mingxian169, enables the identification of Yr27, Yr30, and QYr.nwafu-2BL.4 for adult-plant resistance to stripe rust from Yaco"S" and of Yr18 from Mingxian169. QYr.nwafu-2BL.4 is different from any previously reported gene/QTL. Three haplotypes and six candidate genes have been identified for QYr.nwafu-2BL.4 on the basis of haplotype analysis, micro-collinearity, gene annotation, RNA sequencing, and SNP data. This array provides a new tool for wheat genetic analysis and breeding studies and for achieving durable control of wheat stripe rust.
Keywords: genotyping by target sequencing; liquid array; resistance gene; stripe rust; wheat.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Allard R.W. John Wiley and Sons Inc; 1960. Principles of Plant Breeding.
-
- Allen A.M., Winfield M.O., Burridge A.J., Downie R.C., Benbow H.R., Barker G.L.A., Wilkinson P.A., Coghill J., Waterfall C., Davassi A., et al. Characterization of a wheat breeders array suitable for high-throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum) Plant Biotechnol. J. 2017;15:390–401. - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

