Effective requesting method to detect fusion transcripts in chronic myelomonocytic leukemia RNA-seq
- PMID: 39318504
- PMCID: PMC11420675
- DOI: 10.1093/nargab/lqae117
Effective requesting method to detect fusion transcripts in chronic myelomonocytic leukemia RNA-seq
Abstract
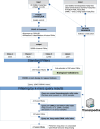
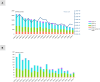
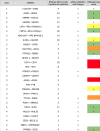
RNA sequencing technology combining short read and long read analysis can be used to detect chimeric RNAs in malignant cells. Here, we propose an integrated approach that uses k-mers to analyze indexed datasets. This approach is used to identify chimeric RNA in chronic myelomonocytic leukemia (CMML) cells, a myeloid malignancy that associates features of myelodysplastic and myeloproliferative neoplasms. In virtually every CMML patient, new generation sequencing identifies one or several somatic driver mutations, typically affecting epigenetic, splicing and signaling genes. In contrast, cytogenetic aberrations are currently detected in only one third of the cases. Nevertheless, chromosomal abnormalities contribute to patient stratification, some of them being associated with higher risk of poor outcome, e.g. through transformation into acute myeloid leukemia (AML). Our approach selects four chimeric RNAs that have been detected and validated in CMML cells. We further focus on NRIP1-MIR99AHG, as this fusion has also recently been detected in AML cells. We show that this fusion encodes three isoforms, including a novel one. Further studies will decipher the biological significance of such a fusion and its potential to improve disease stratification. Taken together, this report demonstrates the ability of a large-scale approach to detect chimeric RNAs in cancer cells.
© The Author(s) 2024. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures




Similar articles
-
Targeted deep sequencing improves outcome stratification in chronic myelomonocytic leukemia with low risk cytogenetic features.Oncotarget. 2016 Aug 30;7(35):57021-57035. doi: 10.18632/oncotarget.10937. Oncotarget. 2016. PMID: 27486981 Free PMC article.
-
Genomic Landscape and Risk Stratification in Chronic Myelomonocytic Leukemia.Curr Hematol Malig Rep. 2021 Jun;16(3):247-255. doi: 10.1007/s11899-021-00613-9. Epub 2021 Mar 3. Curr Hematol Malig Rep. 2021. PMID: 33660195 Review.
-
New chimeric RNAs in acute myeloid leukemia.F1000Res. 2017 Aug 2;6:ISCB Comm J-1302. doi: 10.12688/f1000research.11352.2. eCollection 2017. F1000Res. 2017. PMID: 29623188 Free PMC article.
-
Chronic myelomonocytic leukemia: myeloproliferative variant.Curr Hematol Rep. 2004 May;3(3):218-26. Curr Hematol Rep. 2004. PMID: 15087071 Review.
-
Mutations in chronic myelomonocytic leukemia and their prognostic relevance.Clin Transl Oncol. 2021 Sep;23(9):1731-1742. doi: 10.1007/s12094-021-02585-x. Epub 2021 Apr 16. Clin Transl Oncol. 2021. PMID: 33861431 Review.
References
-
- Arindrarto W., Borràs D.M., de Groen R.A.L., van den Berg R.R., Locher I.J., van Diessen S.A.M.E., van der Holst R., van der Meijden E.D., Honders M.W., de Leeuw R.H.et al. .. Comprehensive diagnostics of acute myeloid leukemia by whole transcriptome RNA sequencing. Leukemia. 2021; 35:47–61. - PMC - PubMed
-
- Shi X., Singh S., Lin E., Li H.. Makowski G.S. Chapter one - Chimeric RNAs in cancer. Advances in Clinical Chemistry. 2021; 100:Elsevier; 1–35. - PubMed
LinkOut - more resources
Full Text Sources

