A review of model evaluation metrics for machine learning in genetics and genomics
- PMID: 39318760
- PMCID: PMC11420621
- DOI: 10.3389/fbinf.2024.1457619
A review of model evaluation metrics for machine learning in genetics and genomics
Abstract
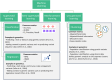
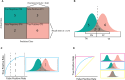
Machine learning (ML) has shown great promise in genetics and genomics where large and complex datasets have the potential to provide insight into many aspects of disease risk, pathogenesis of genetic disorders, and prediction of health and wellbeing. However, with this possibility there is a responsibility to exercise caution against biases and inflation of results that can have harmful unintended impacts. Therefore, researchers must understand the metrics used to evaluate ML models which can influence the critical interpretation of results. In this review we provide an overview of ML metrics for clustering, classification, and regression and highlight the advantages and disadvantages of each. We also detail common pitfalls that occur during model evaluation. Finally, we provide examples of how researchers can assess and utilise the results of ML models, specifically from a genomics perspective.
Keywords: classification; clustering; disease prediction; genomics prediction; machine learning; metrics; regression.
Copyright © 2024 Miller, Portlock, Nyaga and O’Sullivan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Ali M. (2020). PyCaret: an open source, low-code machine learning library in Python. Available at: https://www.pycaret.org.
-
- Babichev S., Lytvynenko M. A. T., Osypenko V. (2017). “Criterial analysis of gene expression sequences to create the objective clustering inductive technology,” in 2017 IEEE 37th international conference on electronics and nanotechnology (ELNANO) (IEEE; ).
Publication types
LinkOut - more resources
Full Text Sources

