[Pathogenesis and potential diagnostic biomarkers of atrial fibrillation in Chinese population: a study based on bioinfor-matics]
- PMID: 39319462
- PMCID: PMC11528137
- DOI: 10.3724/zdxbyxb-2024-0027
[Pathogenesis and potential diagnostic biomarkers of atrial fibrillation in Chinese population: a study based on bioinfor-matics]
Abstract
Objectives: To explore the pathogenesis and potential biomarkers of atrial fibrillation based on bioinformatics.
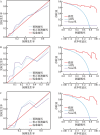
Methods: Differentially expressed genes and module genes related to atrial fibrillation were obtained from GSE41177 and GSE79768 datasets (Chinese-origin tissue samples) through differential expression analysis and weighted gene co-expression network analysis. Candidate hub genes were obtained by taking intersections, and hub genes were obtained after gender stratification. Subsequently, functional enrichment analysis and immune infiltration analysis were performed. Four machine learning models were constructed based on the hub genes, and the optimal model was selected to construct a prediction nomogram. The prediction ability of the nomogram was verified using calibration curves and decision curves. Finally, potential therapeutic drugs for atrial fibrillation were screened from the DGIdb database.
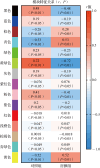
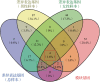
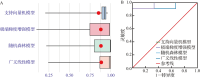
Results: A total of 67 differentially expressed genes and 65 module genes related to atrial fibrillation were identified. Functional enrichment analysis indicated that the pathogenesis of atrial fibrillation was closely related to inflammatory response, immune response, and immune and infectious diseases. Four common hub genes (TYROBP, FCER1G, EVI2B and SOD2), and two genes specifically expressed in male (PILRA and SLC35G3) and female (HLA-DRA and GATP) patients with atrial fibrillation were obtained after gender-segregated screening. The extreme gradient boosting model had satisfactory diagnostic efficiency, and the nomogram constructed based on the hub genes, male significant variables (PILRA, SLC35G3 and SOD2), and female significant variables (FCER1G, SOD2 and TYROBP) had satisfactory predictive ability. Immune infiltration analysis demonstrated a disturbed immune infiltration microenvironment in atrial fibrillation with a higher abundance of plasma cells, neutrophils, and γδT cells, with a higher abundance of neutrophils in males and resting mast cells in females. Two potential drugs for the treatment of atrial fibrillation, valproic acid and methotrexate, were obtained by database and literature screening.
Conclusions: The pathogenesis of atrial fibrillation is closely related to inflammation and immune response, and the microenvironment of immune cell infiltration of cardiomyocytes in the atrial tissue of patients with atrial fibrillation is disordered. TYROBP, FCER1G, EVI2B and SOD2 serve as potential diagnostic biomarkers of atrial fibrillation; PILRA and SLC35G3 serve as potential specific diagnostic biomarkers of atrial fibrillation in the male population, which can effectively predict the risk of atrial fibrillation development and are also potential targets for the treatment of atrial fibrillation.
目的: 基于生物信息学挖掘心房颤动(以下简称房颤)的发病机制和潜在生物标志物。方法: 使用GSE41177和GSE79768数据集进行差异表达分析和加权基因共表达网络分析,获得差异表达基因和模块基因,取交集后获得候选枢纽基因,经性别分层后获得枢纽基因。随后进行功能富集分析和免疫浸润分析。基于枢纽基因构建机器学习模型,选择最优模型构建列线图,并使用校准曲线和决策曲线验证模型的预测能力。最后通过DGIdb数据库筛选治疗房颤的潜在药物。结果: 鉴定出67个差异表达基因和65个与房颤相关的模块基因,功能富集分析表明房颤发病与炎症反应、免疫反应和免疫性、感染性疾病密切相关。经性别分层筛选获得四个具有普适性的枢纽基因(TYROBP、FCER1G、EVI2B、SOD2)、两个在男性房颤患者特异性表达基因(PILRA和SLC35G3)和两个在女性房颤患者特异性表达基因(HLA-DRA和GATP)。极端梯度增强模型的诊断效能优,基于枢纽基因、男性重要基因(PILRA、SLC35G3和SOD2)和女性重要基因(FCER1G、SOD2和TYROBP)所构建的列线图具有较强的预测能力。免疫浸润分析表明,房颤患者的免疫浸润微环境紊乱,具有更高丰富度的浆细胞、中性粒细胞和γδT细胞;男性房颤患者具有更高丰富度的中性粒细胞;女性房颤患者具有更高丰富度的静止肥大细胞。经数据库和文献筛选获得了两种治疗房颤的潜在药物,即丙戊酸和甲氨蝶呤。结论: 房颤的发病机制与炎症和免疫反应密切相关;患者心房组织心肌细胞免疫细胞浸润微环境紊乱;TYROBP、FCER1G、EVI2B和SOD2可作为房颤的潜在诊断生物标志物,PILRA和SLC35G3可作为男性人群房颤的潜在特异性生物标志物,可有效预测房颤的发病风险,也是治疗房颤的潜在靶点。.
Keywords: Atrial fibrillation; Bioinformatics; Biomarkers; Immune response; Inflammatory response; Machine learning models.
Conflict of interest statement
所有作者均声明不存在利益冲突
The authors declare that there is no conflict of interests
Figures
Similar articles
-
PILRA is associated with immune cells infiltration in atrial fibrillation based on bioinformatics and experiment validation.Front Cardiovasc Med. 2023 Jun 16;10:1082015. doi: 10.3389/fcvm.2023.1082015. eCollection 2023. Front Cardiovasc Med. 2023. PMID: 37396579 Free PMC article.
-
Exploring the pathogenesis and immune infiltration in dilated cardiomyopathy complicated with atrial fibrillation by bioinformatics analysis.Front Immunol. 2023 Jan 17;14:1049351. doi: 10.3389/fimmu.2023.1049351. eCollection 2023. Front Immunol. 2023. PMID: 36733486 Free PMC article.
-
Immune-associated pivotal biomarkers identification and competing endogenous RNA network construction in post-operative atrial fibrillation by comprehensive bioinformatics and machine learning strategies.Front Immunol. 2022 Oct 20;13:974935. doi: 10.3389/fimmu.2022.974935. eCollection 2022. Front Immunol. 2022. PMID: 36341343 Free PMC article.
-
CXCR4 and TYROBP mediate the development of atrial fibrillation via inflammation.J Cell Mol Med. 2022 Jun;26(12):3557-3567. doi: 10.1111/jcmm.17405. Epub 2022 May 23. J Cell Mol Med. 2022. PMID: 35607269 Free PMC article.
-
Identification and validation of potential biomarkers for atrial fibrillation based on integrated bioinformatics analysis.Front Cell Dev Biol. 2024 Jan 11;11:1190273. doi: 10.3389/fcell.2023.1190273. eCollection 2023. Front Cell Dev Biol. 2024. PMID: 38274270 Free PMC article.
References
-
- ZHANG J, JOHNSEN S P, GUO Y, et al. . Epidemiology of atrial fibrillation: geographic/ecological risk factors, age, sex, genetics[J]. Card Electrophysiol Clin, 2021, 13(1): 1-23. - PubMed
-
- NATTEL S, HARADA M. Atrial remodeling and atrial fibrillation: recent advances and translational perspec-tives[J]. J Am Coll Cardiol, 2014, 63(22): 2335-2345. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous