Phylogenomic instructed target analysis reveals ELAV complex binding to multiple optimally spaced U-rich motifs
- PMID: 39319593
- PMCID: PMC11551757
- DOI: 10.1093/nar/gkae826
Phylogenomic instructed target analysis reveals ELAV complex binding to multiple optimally spaced U-rich motifs
Abstract
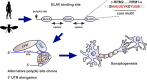
ELAV/Hu RNA-binding proteins are gene-specific regulators of alternative pre-mRNA processing. ELAV/Hu family proteins bind to short AU-rich motifs which are abundant in pre-mRNA, making it unclear how they achieve gene specificity. ELAV/Hu proteins multimerize, but how multimerization contributes to decode degenerate sequence environments remains uncertain. Here, we show that ELAV forms a saturable complex on extended RNA. Through phylogenomic instructed target analysis we identify the core binding motif U5N2U3, which is repeated in an extended binding site. Optimally spaced short U5N2U3 binding motifs are key for high-affinity binding in this minimal binding element. Binding strength correlates with ELAV-regulated alternative poly(A) site choice, which is physiologically relevant through regulation of the major ELAV target ewg in determining synapse numbers. We further identify a stem-loop secondary structure in the ewg binding site unwound upon ELAV binding at three distal U motifs. Base-pairing of U motifs prevents ELAV binding, but N6-methyladenosine (m6A) has little effect. Further, stem-loops are enriched in ELAV-regulated poly(A) sites. Additionally, ELAV can nucleate preferentially from 3' to 5'. Hence, we identify a decisive mechanism for ELAV complex formation, addressing a fundamental gap in understanding how ELAV/Hu family proteins decode degenerate sequence spaces for gene-specific mRNA processing.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures







Similar articles
-
ELAV multimerizes on conserved AU4-6 motifs important for ewg splicing regulation.Mol Cell Biol. 2005 Sep;25(17):7580-91. doi: 10.1128/MCB.25.17.7580-7591.2005. Mol Cell Biol. 2005. PMID: 16107705 Free PMC article.
-
ELAV-mediated 3'-end processing of ewg transcripts is evolutionarily conserved despite sequence degeneration of the ELAV-binding site.Genetics. 2011 Sep;189(1):97-107. doi: 10.1534/genetics.111.131383. Epub 2011 Jul 29. Genetics. 2011. PMID: 21705751 Free PMC article.
-
The Elav-like proteins bind to AU-rich elements and to the poly(A) tail of mRNA.Nucleic Acids Res. 1997 Sep 15;25(18):3564-9. doi: 10.1093/nar/25.18.3564. Nucleic Acids Res. 1997. PMID: 9278474 Free PMC article.
-
Determinants of ELAV gene-specific regulation.Biochem Soc Trans. 2010 Aug;38(4):1122-4. doi: 10.1042/BST0381122. Biochem Soc Trans. 2010. PMID: 20659015 Review.
-
ELAV/Hu RNA-binding protein family: key regulators in neurological disorders, cancer, and other diseases.RNA Biol. 2025 Dec;22(1):1-11. doi: 10.1080/15476286.2025.2471133. Epub 2025 Mar 20. RNA Biol. 2025. PMID: 40000387 Free PMC article. Review.
Cited by
-
Effects of genetic ablation and pharmacological inhibition of HuR on gene expression, iron metabolism, and hormone levels.BMC Biol. 2025 Jan 23;23(1):24. doi: 10.1186/s12915-025-02131-z. BMC Biol. 2025. PMID: 39849491 Free PMC article.
References
-
- Matlin A.J., Clark F., Smith C.W.. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005; 6:386–398. - PubMed
-
- Rogalska M.E., Vivori C., Valcárcel J.. Regulation of pre-mRNA splicing: roles in physiology and disease, and therapeutic prospects. Nat. Rev. Genet. 2023; 24:251–269. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

