Antigenic drift and subtype interference shape A(H3N2) epidemic dynamics in the United States
- PMID: 39319780
- PMCID: PMC11424097
- DOI: 10.7554/eLife.91849
Antigenic drift and subtype interference shape A(H3N2) epidemic dynamics in the United States
Abstract
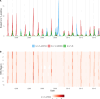
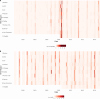
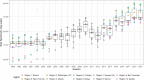
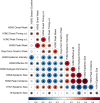
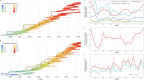
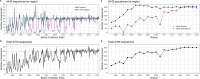
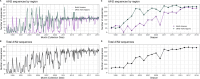
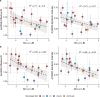
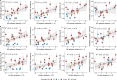
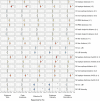
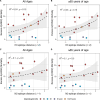
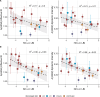
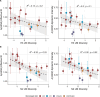
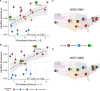
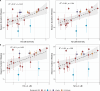
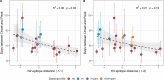
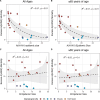
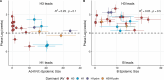
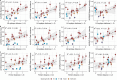
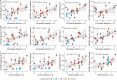
Influenza viruses continually evolve new antigenic variants, through mutations in epitopes of their major surface proteins, hemagglutinin (HA) and neuraminidase (NA). Antigenic drift potentiates the reinfection of previously infected individuals, but the contribution of this process to variability in annual epidemics is not well understood. Here, we link influenza A(H3N2) virus evolution to regional epidemic dynamics in the United States during 1997-2019. We integrate phenotypic measures of HA antigenic drift and sequence-based measures of HA and NA fitness to infer antigenic and genetic distances between viruses circulating in successive seasons. We estimate the magnitude, severity, timing, transmission rate, age-specific patterns, and subtype dominance of each regional outbreak and find that genetic distance based on broad sets of epitope sites is the strongest evolutionary predictor of A(H3N2) virus epidemiology. Increased HA and NA epitope distance between seasons correlates with larger, more intense epidemics, higher transmission, greater A(H3N2) subtype dominance, and a greater proportion of cases in adults relative to children, consistent with increased population susceptibility. Based on random forest models, A(H1N1) incidence impacts A(H3N2) epidemics to a greater extent than viral evolution, suggesting that subtype interference is a major driver of influenza A virus infection ynamics, presumably via heterosubtypic cross-immunity.
Keywords: H3N2; antigenic drift; epidemiology; global health; human; infectious disease; influenza virus; microbiology; virus.
Plain language summary
Seasonal influenza (flu) viruses cause outbreaks every winter. People infected with influenza typically develop mild respiratory symptoms. But flu infections can cause serious illness in young children, older adults and people with chronic medical conditions. Infected or vaccinated individuals develop some immunity, but the viruses evolve quickly to evade these defenses in a process called antigenic drift. As the viruses change, they can re-infect previously immune people. Scientists update the flu vaccine yearly to keep up with this antigenic drift. The immune system fights flu infections by recognizing two proteins, known as antigens, on the virus’s surface, called hemagglutinin (HA) and neuraminidase (NA). However, mutations in the genes encoding these proteins can make them unrecognizable, letting the virus slip past the immune system. Scientists would like to know how these changes affect the size, severity and timing of annual influenza outbreaks. Perofsky et al. show that tracking genetic changes in HA and NA may help improve flu season predictions. The experiments compared the severity of 22 flu seasons caused by the A(H3N2) subtype in the United States with how much HA and NA had evolved since the previous year. The A(H3N2) subtype experiences the fastest rates of antigenic drift and causes more cases and deaths than other seasonal flu viruses. Genetic changes in HA and NA were a better predictor of A(H3N2) outbreak severity than the blood tests for protective antibodies that epidemiologists traditionally use to track flu evolution. However, the prevalence of another subtype of influenza A circulating in the population, called A(H1N1), was an even better predictor of how severe A(H3N2) outbreaks would be. Perofsky et al. are the first to show that genetic changes in NA contribute to the severity of flu seasons. Previous studies suggested a link between genetic changes in HA and flu season severity, and flu vaccines include the HA protein to help the body recognize new influenza strains. The results suggest that adding the NA protein to flu vaccines may improve their effectiveness. In the future, flu forecasters may want to analyze genetic changes in both NA and HA to make their outbreak predictions. Tracking how much of the A(H1N1) subtype is circulating may also be useful for predicting the severity of A(H3N2) outbreaks.
Conflict of interest statement
AP, JH, JB, TR, XX, RK, DW, NL, LW, BE, RH, MG, RD, SF, KN, NK, SW, HH, TB No competing interests declared, CH Received personal fees from Sanofi outside the submitted work, JM Received consulting fees, honoraria, and travel support from Sanofi Pasteur and Sequris, SS The WHO Collaborating Centre for Reference and Research on Influenza in Melbourne has a collaborative research and development agreement (CRADA) with CSL Seqirus for isolation of candidate vaccine viruses in cells and an agreement with IFPMA for isolation of candidate vaccine viruses in eggs. SGS reports honoraria from CSL Seqirus, Moderna, Pfizer, and Evo Health, IB, KS The WHO Collaborating Centre for Reference and Research on Influenza in Melbourne has a collaborative research and development agreement (CRADA) with CSL Seqirus for isolation of candidate vaccine viruses in cells and an agreement with IFPMA for isolation of candidate vaccine viruses in eggs, FK The Icahn School of Medicine at Mount Sinai has filed patent applications relating to influenza virus vaccines (U.S. patent numbers: 12030928, 11865173, 11266734, 11254733, 10736956, 10583188, 10137189, 10131695, 9968670, 9371366; publication numbers: 20230181715, 20220403358, 20220249652, 20220242935, 20220153873, 20210260179, 20190125859, 20190106461, 20180333479), SARS-CoV-2 serological assays (publication number: 20240210415), and SARS-CoV-2 vaccines (publication numbers: 20230310583, 20230226171), which list FK as co-inventor. FK has consulted for Merck and Pfizer (before 2020), and is currently consulting for Pfizer, Seqirus, 3rd Rock Ventures, GSK and Avimex. The Krammer laboratory is also collaborating with Pfizer on animal models of SARS‐CoV‐2 and with Dynavax on universal influenza virus vaccines, CV Received honoraria for serving as an Editor in Chief of the journal Epidemics (Elsevier)
Figures
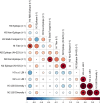
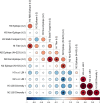
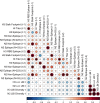
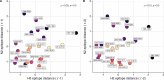


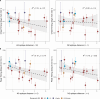
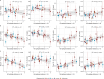
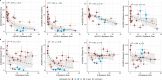






Update of
-
Antigenic drift and subtype interference shape A(H3N2) epidemic dynamics in the United States.medRxiv [Preprint]. 2024 May 22:2023.10.02.23296453. doi: 10.1101/2023.10.02.23296453. medRxiv. 2024. Update in: Elife. 2024 Sep 25;13:RP91849. doi: 10.7554/eLife.91849. PMID: 37873362 Free PMC article. Updated. Preprint.
References
-
- Ali ST, Lau YC, Shan S, Ryu S, Du Z, Wang L, Xu XK, Chen D, Xiong J, Tae J, Tsang TK, Wu P, Lau EHY, Cowling BJ. Prediction of upcoming global infection burden of influenza seasons after relaxation of public health and social measures during the COVID-19 pandemic: a modelling study. The Lancet. Global Health. 2022;10:e1612–e1622. doi: 10.1016/S2214-109X(22)00358-8. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 10111800/Ministry of Health, Labour and Welfare
- 75N93019C00051/NH/NIH HHS/United States
- FC001030/WT_/Wellcome Trust/United Kingdom
- 10110400/Ministry of Health, Labour and Welfare
- JP22fk0108118/Japan Agency for Medical Research and Development
- HHSN272201400008C/NH/NIH HHS/United States
- F31 AI140714/NH/NIH HHS/United States
- 75N93021C00014/AI/NIAID NIH HHS/United States
- 75N93019C00051/AI/NIAID NIH HHS/United States
- 75N93021C00014/NH/NIH HHS/United States
- R01 AI165821/AI/NIAID NIH HHS/United States
- R01 AI165821/NH/NIH HHS/United States
- HHSN272201400008C/AI/NIAID NIH HHS/United States
- F31 AI140714/AI/NIAID NIH HHS/United States
- 1354890/National Science Foundation
- R35 GM119774/GM/NIGMS NIH HHS/United States
- R35 GM119774/NH/NIH HHS/United States
- R01 AI127893/NH/NIH HHS/United States
- FC001030/MRC_/Medical Research Council/United Kingdom
- JP23fk0108662/Japan Agency for Medical Research and Development
- FC001030/CRUK_/Cancer Research UK/United Kingdom
- R01 AI127893/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

