Sialic acid and PirB are not required for targeting of neural circuits by neurotropic mammalian orthoreovirus
- PMID: 39320067
- PMCID: PMC11540169
- DOI: 10.1128/msphere.00629-24
Sialic acid and PirB are not required for targeting of neural circuits by neurotropic mammalian orthoreovirus
Abstract
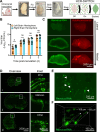
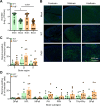
Serotype 3 (T3) strains of mammalian orthoreovirus (reovirus) spread to the central nervous system to infect the brain and cause lethal encephalitis in newborn mice. Although reovirus targets several regions in the brain, susceptibility to infection is not uniformly distributed. The neuronal subtypes and anatomic sites targeted throughout the brain are not precisely known. Reovirus binds several attachment factors and entry receptors, including sialic acid (SA)-containing glycans and paired immunoglobulin-like receptor B (PirB). While these receptors are not required for infection of some types of neurons, reovirus engagement of these receptors can influence neuronal infection in certain contexts. To identify patterns of T3 neurotropism, we used microbial identification after passive tissue clearance and hybridization chain reaction to stain reovirus-infected cells throughout intact, optically transparent brains of newborn mice. Three-dimensional reconstructions revealed in detail the sites targeted by reovirus throughout the brain volume, including dense infection of the midbrain and hindbrain. Using reovirus mutants incapable of binding SA and mice lacking PirB expression, we found that neither SA nor PirB is required for the infection of various brain regions. However, SA may confer minor differences in infection that vary by region. Collectively, these studies indicate that many regions in the brain of newborn mice are susceptible to reovirus and that patterns of reovirus infection are not dependent on reovirus receptors SA and PirB.IMPORTANCENeurotropic viruses invade the central nervous system (CNS) and target various cell types to cause disease manifestations, such as meningitis, myelitis, or encephalitis. Infections of the CNS are often difficult to treat and can lead to lasting sequelae or death. Mammalian orthoreovirus (reovirus) causes age-dependent lethal encephalitis in many young mammals. Reovirus infects neurons in several different regions of the brain. However, the complete pattern of CNS infection is not understood. We found that reovirus targets almost all regions of the brain and that patterns of tropism are not dependent on receptors sialic acid and paired immunoglobulin-like receptor B. These studies confirm that two known reovirus receptors do not completely explain the cell types infected in brain tissue and establish strategies that can be used to understand complete patterns of viral tropism in an intact brain.
Keywords: CLARITY; MiPACT-HCR; PirB; neuropathogenesis; neurotropic viruses; reovirus; sialic acid.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Paired immunoglobulin-like receptor B is an entry receptor for mammalian orthoreovirus.Nat Commun. 2023 May 5;14(1):2615. doi: 10.1038/s41467-023-38327-6. Nat Commun. 2023. PMID: 37147336 Free PMC article.
-
Reovirus Neurotropism and Virulence Are Dictated by Sequences in the Head Domain of the Viral Attachment Protein.J Virol. 2018 Nov 12;92(23):e00974-18. doi: 10.1128/JVI.00974-18. Print 2018 Dec 1. J Virol. 2018. PMID: 30209169 Free PMC article.
-
Strain-specific differences in reovirus infection of murine macrophages segregate with polymorphisms in viral outer-capsid protein σ3.J Virol. 2024 Nov 19;98(11):e0114724. doi: 10.1128/jvi.01147-24. Epub 2024 Oct 21. J Virol. 2024. PMID: 39431846 Free PMC article.
-
An Orchestra of Reovirus Receptors: Still Searching for the Conductor.Adv Virus Res. 2018;100:223-246. doi: 10.1016/bs.aivir.2017.10.005. Epub 2017 Nov 13. Adv Virus Res. 2018. PMID: 29551138 Review.
-
Attachment and cell entry of mammalian orthoreovirus.Curr Top Microbiol Immunol. 2006;309:1-38. doi: 10.1007/3-540-30773-7_1. Curr Top Microbiol Immunol. 2006. PMID: 16909895 Review.
References
-
- Dermody TS, Parker JS, Sherry B. 2023. Orthoreoviruses, p 318–361. In Howley PM, Knip DM (ed), Fields virology, 7th ed. Wolters Kluwer, Philadelphia.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

