MmpL3, Wag31, and PlrA are involved in coordinating polar growth with peptidoglycan metabolism and nutrient availability
- PMID: 39320104
- PMCID: PMC11500546
- DOI: 10.1128/jb.00204-24
MmpL3, Wag31, and PlrA are involved in coordinating polar growth with peptidoglycan metabolism and nutrient availability
Abstract
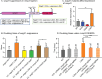
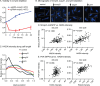
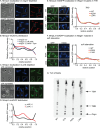
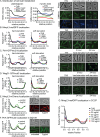
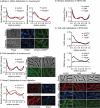
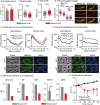
Cell growth in mycobacteria involves cell wall expansion that is restricted to the cell poles. The DivIVA homolog Wag31 is required for this process, but the molecular mechanism and protein partners of Wag31 have not been described. In this study of Mycobacterium smegmatis, we identify a connection between wag31 and trehalose monomycolate (TMM) transporter mmpl3 in a suppressor screen and show that Wag31 and polar regulator PlrA are required for MmpL3's polar localization. In addition, the localization of PlrA and MmpL3 is responsive to nutrient and energy deprivation and inhibition of peptidoglycan metabolism. We show that inhibition of MmpL3 causes delocalized cell wall metabolism but does not delocalize MmpL3 itself. We found that cells with an MmpL3 C-terminal truncation, which is defective for localization, have only minor defects in polar growth but are impaired in their ability to downregulate cell wall metabolism under stress. Our work suggests that, in addition to its established function in TMM transport, MmpL3 has a second function in regulating global cell wall metabolism in response to stress. Our data are consistent with a model in which the presence of TMMs in the periplasm stimulates polar elongation and in which the connection between Wag31, PlrA, and the C-terminus of MmpL3 is involved in detecting and responding to stress in order to coordinate the synthesis of the different layers of the mycobacterial cell wall in changing conditions.
Importance: This study is performed in Mycobacterium smegmatis, which is used as a model to understand the basic physiology of pathogenic mycobacteria such as Mycobacterium tuberculosis. In this work, we examine the function and regulation of three proteins involved in regulating cell wall elongation in mycobacterial cells, which occurs at the cell tips or poles. We find that Wag31, a regulator of polar elongation, works partly through the regulation of MmpL3, a transporter of cell wall constituents and an important drug target. Our work suggests that, beyond its transport function, MmpL3 has another function in controlling cell wall synthesis broadly in response to stress.
Keywords: DivIVA; MmpL3; Mycobacterium; PlrA; TMMs; murJ; mycolic acid; peptidoglycan; polar growth regulation; stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Update of
-
MmpL3, Wag31 and PlrA are involved in coordinating polar growth with peptidoglycan metabolism and nutrient availability.bioRxiv [Preprint]. 2024 Apr 30:2024.04.29.591792. doi: 10.1101/2024.04.29.591792. bioRxiv. 2024. Update in: J Bacteriol. 2024 Oct 24;206(10):e0020424. doi: 10.1128/jb.00204-24. PMID: 38746181 Free PMC article. Updated. Preprint.
References
-
- Daffé M, Draper P. 1997. The envelope layers of Mycobacteria with reference to their pathogenicity, p 131–203. In Advances in Microbial Physiology. Elsevier. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

