The G Protein-Coupled Receptor GPR56 Is an Inhibitory Checkpoint for NK Cell Migration
- PMID: 39320215
- PMCID: PMC11491499
- DOI: 10.4049/jimmunol.2400228
The G Protein-Coupled Receptor GPR56 Is an Inhibitory Checkpoint for NK Cell Migration
Abstract
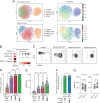
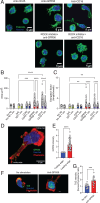
G protein-coupled receptors (GPCRs) represent the largest family of surface receptors and are responsible for key physiological functions, including cell growth, neurotransmission, hormone release, and cell migration. The GPCR 56 (GPR56), encoded by ADGRG1, is an adhesion GPCR found on diverse cell types, including neural progenitor cells, melanoma cells, and lymphocytes, such as effector memory T cells, γδ T cells, and NK cells. Using RNA-sequencing and high-resolution flow cytometry, we found that GPR56 mRNA and protein expression increased with NK cell differentiation, reaching its peak in adaptive NK cells. Small interfering RNA silencing of GPR56 led to increased spontaneous and chemokine-induced migration, suggesting that GPR56 functions as an upstream checkpoint for migration of highly differentiated NK cells. Increased NK cell migration could also be induced by agonistic stimulation of GPR56 leading to rapid internalization and deactivation of the receptor. Mechanistically, GPR56 ligation and downregulation were associated with transcriptional coactivator with PDZ-binding motif translocation to the nucleus and increased actin polymerization. Together, these data provide insights into the role of GPR56 in the migratory behavior of human NK cell subsets and may open possibilities to improve NK cell infiltration into cancer tissues by releasing a migratory checkpoint.
Copyright © 2024 by The American Association of Immunologists, Inc.
Conflict of interest statement
K.-J.M. is a consultant for and has received research support for unrelated studies from Fate Therapeutics, has received research support from Oncopeptides, and is a consultant at Vycellix. All relationships have been approved by Oslo University Hospital, University of Oslo, and Karolinska Institute. The other authors have no financial conflicts of interest.
Figures


References
-
- Walzer, T., Vivier E.. 2011. G-protein-coupled receptors in control of natural killer cell migration. Trends Immunol. 32: 486–492. - PubMed
-
- Ganesh, R. A., Venkataraman K., Sirdeshmukh R.. 2020. GPR56: an adhesion GPCR involved in brain development, neurological disorders and cancer. Brain Res. 1747: 147055. - PubMed
MeSH terms
Substances
Grants and funding
- 2021-073/Ministry of Health and Care Services | Helse Sør-Øst RHF (sorost)
- 332727/Norges Forskningsråd (Forskningsrådet)
- 237579/Norges Forskningsråd (Forskningsrådet)
- 2024053/Ministry of Health and Care Services | Helse Sør-Øst RHF (sorost)
- 223310/Kreftforeningen (NCS)
- 2021-073/Ministry of Health and Care Services | Helse Sør-Øst RHF (sorost)
- PR2022-0150/Barncancerfonden (Swedish Childhood Cancer Foundation)
- Knut och Alice Wallenbergs Stiftelse (Knut and Alice Wallenberg Foundation)
- 2024053/Ministry of Health and Care Services | Helse Sør-Øst RHF (sorost)
- 275469/Norges Forskningsråd (Forskningsrådet)
- 801133/EC | Horizon Europe | Excellent Science | HORIZON EUROPE Marie Sklodowska-Curie Actions (MSCA)
- 211793/Cancerfonden (Swedish Cancer Society)
- P01 CA111412/CA/NCI NIH HHS/United States
- 332727/Norges Forskningsråd (Forskningsrådet)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

