Explainable breast cancer molecular expression prediction using multi-task deep-learning based on 3D whole breast ultrasound
- PMID: 39320560
- PMCID: PMC11424596
- DOI: 10.1186/s13244-024-01810-9
Explainable breast cancer molecular expression prediction using multi-task deep-learning based on 3D whole breast ultrasound
Abstract
Objectives: To noninvasively estimate three breast cancer biomarkers, estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) and enhance performance and interpretability via multi-task deep learning.
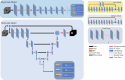
Methods: The study included 388 breast cancer patients who received the 3D whole breast ultrasound system (3DWBUS) examinations at Xijing Hospital between October 2020 and September 2021. Two predictive models, a single-task and a multi-task, were developed; the former predicts biomarker expression, while the latter combines tumor segmentation with biomarker prediction to enhance interpretability. Performance evaluation included individual and overall prediction metrics, and Delong's test was used for performance comparison. The models' attention regions were visualized using Grad-CAM + + technology.
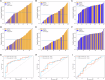
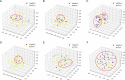
Results: All patients were randomly split into a training set (n = 240, 62%), a validation set (n = 60, 15%), and a test set (n = 88, 23%). In the individual evaluation of ER, PR, and HER2 expression prediction, the single-task and multi-task models achieved respective AUCs of 0.809 and 0.735 for ER, 0.688 and 0.767 for PR, and 0.626 and 0.697 for HER2, as observed in the test set. In the overall evaluation, the multi-task model demonstrated superior performance in the test set, achieving a higher macro AUC of 0.733, in contrast to 0.708 for the single-task model. The Grad-CAM + + method revealed that the multi-task model exhibited a stronger focus on diseased tissue areas, improving the interpretability of how the model worked.
Conclusion: Both models demonstrated impressive performance, with the multi-task model excelling in accuracy and offering improved interpretability on noninvasive 3DWBUS images using Grad-CAM + + technology.
Critical relevance statement: The multi-task deep learning model exhibits effective prediction for breast cancer biomarkers, offering direct biomarker identification and improved clinical interpretability, potentially boosting the efficiency of targeted drug screening.
Key points: Tumoral biomarkers are paramount for determining breast cancer treatment. The multi-task model can improve prediction performance, and improve interpretability in clinical practice. The 3D whole breast ultrasound system-based deep learning models excelled in predicting breast cancer biomarkers.
Keywords: Breast cancer; Deep learning; Ultrasound imaging.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Zhang T, Tan T, Samperna R et al (2023) Radiomics and artificial intelligence in breast imaging: a survey. Artif Intell Rev. 10.1007/s10462-023-10543-y
Grants and funding
- 20220810144949003/Shenzhen Natural Science Fund (the Stable Support Plan Program)
- 20220810144949003/Shenzhen Natural Science Fund (the Stable Support Plan Program)
- 20220810144949003/Shenzhen Natural Science Fund (the Stable Support Plan Program)
- 20220810144949003/Shenzhen Natural Science Fund (the Stable Support Plan Program)
- 20220810144949003/Shenzhen Natural Science Fund (the Stable Support Plan Program)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

