Emerging SARS-CoV-2 Resistance After Antiviral Treatment
- PMID: 39320890
- PMCID: PMC11425144
- DOI: 10.1001/jamanetworkopen.2024.35431
Emerging SARS-CoV-2 Resistance After Antiviral Treatment
Abstract
Importance: Previous studies have identified mutations in SARS-CoV-2 strains that confer resistance to nirmatrelvir, yet how often this resistance arises and its association with posttreatment virologic rebound is not well understood.
Objective: To examine the prevalence of emergent antiviral resistance after nirmatrelvir treatment and its association with virologic rebound.
Design, setting, and participants: This cohort study enrolled outpatient adults with acute COVID-19 infection from May 2021 to October 2023. Participants were divided into those who received antiviral therapy and those who did not. The study was conducted at a multicenter health care system in Boston, Massachusetts.
Exposure: Treatment regimen, including none, nirmatrelvir, and remdesivir.
Main outcomes and measures: The primary outcome was emergent SARS-CoV-2 antiviral resistance, defined as the detection of antiviral resistance mutations, which were not present at baseline, were previously associated with decreased antiviral efficacy, and emerged during or after completion of a participant's treatment. Next-generation sequencing was used to detect low frequency mutations down to 1% of the total viral population.
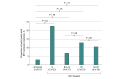
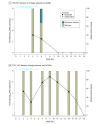
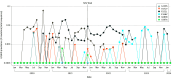
Results: Overall, 156 participants (114 female [73.1%]; median [IQR] age, 56 [38-69] years) were included. Compared with 63 untreated individuals, the 79 who received nirmatrelvir were older and more commonly immunosuppressed. After sequencing viral RNA from participants' anterior nasal swabs, nirmatrelvir resistance mutations were detected in 9 individuals who received nirmatrelvir (11.4%) compared with 2 of those who did not (3.2%) (P = .09). Among the individuals treated with nirmatrelvir, those who were immunosuppressed had the highest frequency of resistance emergence (5 of 22 [22.7%]), significantly greater than untreated individuals (2 of 63 [3.1%]) (P = .01). Similar rates of nirmatrelvir resistance were found in those who had virologic rebound (3 of 23 [13.0%]) vs those who did not (6 of 56 [10.7%]) (P = .86). Most of these mutations (10 of 11 [90.9%]) were detected at low frequencies (<20% of viral population) and reverted to the wild type at subsequent time points. Emerging remdesivir resistance mutations were only detected in immunosuppressed individuals (2 of 14 [14.3%]) but were similarly low frequency and transient. Global Initiative on Sharing All Influenza Data analysis showed no evidence of increased nirmatrelvir resistance in the United States after the authorization of nirmatrelvir.
Conclusions and relevance: In this cohort study of 156 participants, treatment-emergent nirmatrelvir resistance mutations were commonly detected, especially in individuals who were immunosuppressed. However, these mutations were generally present at low frequencies and were transient in nature, suggesting a low risk for the spread of nirmatrelvir resistance in the community with the current variants and drug usage patterns.
Conflict of interest statement
Figures
Comment in
-
SARS-CoV-2 Nirmatrelvir Resistance-A Concern for Immunocompromised Populations?JAMA Netw Open. 2024 Sep 3;7(9):e2435439. doi: 10.1001/jamanetworkopen.2024.35439. JAMA Netw Open. 2024. PMID: 39320895 No abstract available.
References
-
- US Food and Drug Administration. Fact sheet for healthcare providers: emergency use authorization for Paxlovid. Accessed August 21, 2024. https://www.fda.gov/media/155050/download
-
- US Food and Drug Administration. Highlights of prescribing information: Veklury. Accessed August 21, 2024. https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/214787Orig1s00...
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

