Spermidine metabolism regulates leukemia stem and progenitor cell function through KAT7 expression in patient-derived mouse models
- PMID: 39321266
- PMCID: PMC11670652
- DOI: 10.1126/scitranslmed.adn1285
Spermidine metabolism regulates leukemia stem and progenitor cell function through KAT7 expression in patient-derived mouse models
Abstract
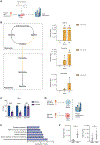
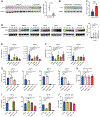
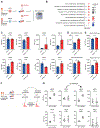
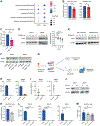
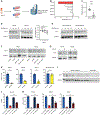
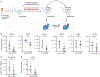
Acute myeloid leukemia (AML) is a devastating disease initiated and maintained by a rare subset of cells called leukemia stem cells (LSCs). LSCs are responsible for driving disease relapse, making the development of new therapeutic strategies to target LSCs urgently needed. The use of mass spectrometry-based metabolomics profiling has enabled the discovery of unique and targetable metabolic properties in LSCs. However, we do not have a comprehensive understanding of metabolite differences between LSCs and their normal counterparts, hematopoietic stem and progenitor cells (HSPCs). In this study, we used an unbiased mass spectrometry-based metabolomics analysis to define differences in metabolites between primary human LSCs and HSPCs, which revealed that LSCs have a distinct metabolome. Spermidine was the most enriched metabolite in LSCs compared with HSPCs. Pharmacological reduction of spermidine concentrations decreased LSC function but spared normal HSPCs. Polyamine depletion also decreased leukemic burden in patient-derived xenografts. Mechanistically, spermidine depletion induced LSC myeloid differentiation by decreasing eIF5A-dependent protein synthesis, resulting in reduced expression of a select subset of proteins. KAT7, a histone acetyltransferase, was one of the top candidates identified to be down-regulated by spermidine depletion. Overexpression of KAT7 partially rescued polyamine depletion-induced decreased colony-forming ability, demonstrating that loss of KAT7 is an essential part of the mechanism by which spermidine depletion targets AML clonogenic potential. Together, we identified and mechanistically dissected a metabolic vulnerability of LSCs that has the potential to be rapidly translated into clinical trials to improve outcomes for patients with AML.
Conflict of interest statement
Figures


References
-
- DiNardo CD, Erba HP, Freeman SD, Wei AH, Acute myeloid leukaemia. Lancet 401, 2073–2086 (2023). - PubMed
-
- Shlush LI, Mitchell A, Heisler L, Abelson S, Ng SWK, Trotman-Grant A, Medeiros JJF, Rao-Bhatia A, Jaciw-Zurakowsky I, Marke R, McLeod JL, Doedens M, Bader G, Voisin V, Xu C, McPherson JD, Hudson TJ, Wang JCY, Minden MD, Dick JE, Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature 547, 104–108 (2017). - PubMed
-
- Hanahan D, Hallmarks of cancer: New dimensions. Cancer Discov. 12, 31–46 (2022). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

