RCC1 regulation of subcellular protein localization via Ran GTPase drives pancreatic ductal adenocarcinoma growth
- PMID: 39321913
- PMCID: PMC11471368
- DOI: 10.1016/j.canlet.2024.217275
RCC1 regulation of subcellular protein localization via Ran GTPase drives pancreatic ductal adenocarcinoma growth
Abstract
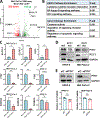
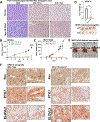
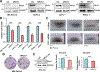
Pancreatic ductal adenocarcinoma (PDAC) is a highly lethal malignancy, with limited therapeutic options. Here, we evaluated the role of regulator of chromosome condensation 1 (RCC1) in PDAC. RCC1 functions as a guanine exchange factor for GTP-binding nuclear protein Ran (Ran) GTPase and is involved in nucleocytoplasmic transport. RCC1 RNA expression is elevated in PDAC tissues compared to normal pancreatic tissues and correlates with poor prognosis. RCC1 silencing by RNAi and CRISPR-Cas9 knockout (KO) results in reduced proliferation in 2-D and 3-D cell cultures. RCC1 knockdown (KD) reduced migration and clonogenicity, enhanced apoptosis, and altered cell cycle progression in human PDAC and murine cells from LSL-KrasG12D/+; LSL-Trp53R172H/+; Pdx1-Cre (KPC) tumors. Mechanistically, RCC1 KO shows widespread transcriptomic alterations including regulation of PTK7, a co-receptor of the Wnt signaling pathway. RCC1 KD disrupted subcellular Ran localization and the Ran gradient. Nuclear and cytosolic proteomics revealed altered subcellular proteome localization in Rcc1 KD KPC-tumor-derived cells and several altered metabolic biosynthesis pathways. In vivo, RCC1 KO cells show reduced tumor growth potential when injected as sub-cutaneous xenografts. Finally, RCC1 KD sensitized PDAC cells to gemcitabine chemotherapy treatment. This study reveals the role of RCC1 in pancreatic cancer as a novel molecular vulnerability that could be exploited to enhance therapeutic response.
Keywords: Nuclear export; PDAC; Pancreatic cancer; RCC1; Ran GTPase.
Copyright © 2024 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Boris C Pasche reports a relationship with Merck & Co Inc that includes: funding grants. Boris C Pasche reports a relationship with Roche that includes: funding grants. Boris C Pasche reports a relationship with Novartis that includes: funding grants. Boris C Pasche reports a relationship with AstraZeneca that includes: funding grants. Boris C Pasche reports a relationship with Bristol Myers Squibb Co that includes: funding grants. Boris C Pasche reports a relationship with TheraBionic Inc that includes: equity or stocks. Boris C Pasche reports a relationship with TheraBionic GmbH that includes: equity or stocks. Asfar S Azmi reports a relationship with Guidepoint that includes: consulting or advisory. Asfar S Azmi reports a relationship with GLG that includes: consulting or advisory. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
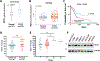


References
-
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48. - PubMed
-
- Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53(4):549–554. - PubMed
-
- Seki T, Hayashi N, Nishimoto T. RCC1 in the Ran pathway. Journal of biochemistry. 1996;120(2):207–214. - PubMed
-
- Bischoff FR, Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354(6348):80–82. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

