Designed endocytosis-inducing proteins degrade targets and amplify signals
- PMID: 39322662
- PMCID: PMC11839401
- DOI: 10.1038/s41586-024-07948-2
Designed endocytosis-inducing proteins degrade targets and amplify signals
Abstract
Endocytosis and lysosomal trafficking of cell surface receptors can be triggered by endogenous ligands. Therapeutic approaches such as lysosome-targeting chimaeras1,2 (LYTACs) and cytokine receptor-targeting chimeras3 (KineTACs) have used this to target specific proteins for degradation by fusing modified native ligands to target binding proteins. Although powerful, these approaches can be limited by competition with native ligands and requirements for chemical modification that limit genetic encodability and can complicate manufacturing, and, more generally, there may be no native ligands that stimulate endocytosis through a given receptor. Here we describe computational design approaches for endocytosis-triggering binding proteins (EndoTags) that overcome these challenges. We present EndoTags for insulin-like growth factor 2 receptor (IGF2R) and asialoglycoprotein receptor (ASGPR), sortilin and transferrin receptors, and show that fusing these tags to soluble or transmembrane target protein binders leads to lysosomal trafficking and target degradation. As these receptors have different tissue distributions, the different EndoTags could enable targeting of degradation to different tissues. EndoTag fusion to a PD-L1 antibody considerably increases efficacy in a mouse tumour model compared to antibody alone. The modularity and genetic encodability of EndoTags enables AND gate control for higher-specificity targeted degradation, and the localized secretion of degraders from engineered cells. By promoting endocytosis, EndoTag fusion increases signalling through an engineered ligand-receptor system by nearly 100-fold. EndoTags have considerable therapeutic potential as targeted degradation inducers, signalling activators for endocytosis-dependent pathways, and cellular uptake inducers for targeted antibody-drug and antibody-RNA conjugates.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: B.H., M. Abedi, G.A., I.S., L.S. and D.B. are co-inventors on a provisional patent application that incorporates discoveries described in this manuscript. B.C., I.G., J.O., P.G., L.S. and D.B. are co-inventors on a provisional patent application that incorporates discoveries described in this manuscript.
Figures
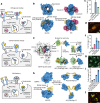

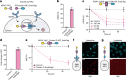
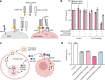




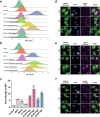




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

