Kinesin-1 mediates proper ER folding of the CaV1.2 channel and maintains mouse glucose homeostasis
- PMID: 39322740
- PMCID: PMC11549326
- DOI: 10.1038/s44319-024-00246-y
Kinesin-1 mediates proper ER folding of the CaV1.2 channel and maintains mouse glucose homeostasis
Abstract
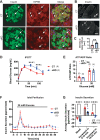
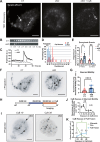
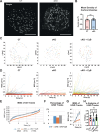
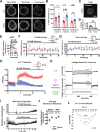
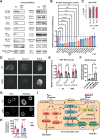
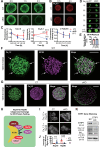
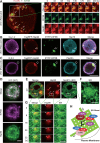
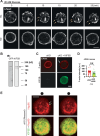
Glucose-stimulated insulin secretion (GSIS) from pancreatic beta cells is a principal mechanism for systemic glucose homeostasis, of which regulatory mechanisms are still unclear. Here we show that kinesin molecular motor KIF5B is essential for GSIS through maintaining the voltage-gated calcium channel CaV1.2 levels, by facilitating an Hsp70-to-Hsp90 chaperone exchange to pass through the quality control in the endoplasmic reticulum (ER). Phenotypic analyses of KIF5B conditional knockout (cKO) mouse beta cells revealed significant abolishment of glucose-stimulated calcium transients, which altered the behaviors of insulin granules via abnormally stabilized cortical F-actin. KIF5B and Hsp90 colocalize to microdroplets on ER sheets, where CaV1.2 but not Kir6.2 is accumulated. In the absence of KIF5B, CaV1.2 fails to be transferred from Hsp70 to Hsp90 via STIP1, and is likely degraded via the proteasomal pathway. KIF5B and Hsc70 overexpression increased CaV1.2 expression via enhancing its chaperone binding. Thus, ER sheets may serve as the place of KIF5B- and Hsp90-dependent chaperone exchange, which predominantly facilitates CaV1.2 production in beta cells and properly enterprises GSIS against diabetes.
Keywords: Calcium Channel; ER Sheets; Hsp90; Insulin Secretion; Kinesin.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Abramoff MD, Magelhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics Int 11:36–42
-
- Altier C, Garcia-Caballero A, Simms B, You H, Chen L, Walcher J, Tedford HW, Hermosilla T, Zamponi GW (2011) The Cavbeta subunit prevents RFP2-mediated ubiquitination and proteasomal degradation of L-type channels. Nat Neurosci 14:173–180 - PubMed
-
- Aoki R, Kitaguchi T, Oya M, Yanagihara Y, Sato M, Miyawaki A, Tsuboi T (2010) Duration of fusion pore opening and the amount of hormone released are regulated by myosin II during kiss-and-run exocytosis. Biochem J 429:497–504 - PubMed
-
- Arous C, Halban PA (2015) The skeleton in the closet: actin cytoskeletal remodeling in beta-cell function. Am J Physiol Endocrinol Metab 309:E611–E620 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

