Increasing intracellular dNTP levels improves prime editing efficiency
- PMID: 39322763
- PMCID: PMC12092096
- DOI: 10.1038/s41587-024-02405-x
Increasing intracellular dNTP levels improves prime editing efficiency
Abstract
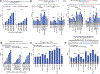
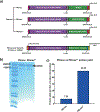
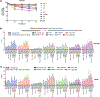
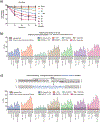
In primary cell types, intracellular deoxynucleotide triphosphate (dNTP) levels are tightly regulated in a cell cycle-dependent manner. We report that prime editing efficiency is increased by mutations that improve the enzymatic properties of Moloney murine leukemia virus reverse transcriptase and treatments that increase intracellular dNTP levels. In combination, these modifications produce substantial increases in precise editing rates.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: E.J.S. is a cofounder and Scientific Advisory Board member of Intellia Therapeutics and a Scientific Advisory Board member at Tessera Therapeutics. S.A.W. is a consultant for Editas Medicine. The University of Massachusetts Chan Medical School has filed patent applications related to this work. All other authors have no competing interests.
Figures

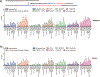
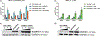
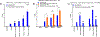
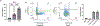

Update of
-
Addressing the dNTP bottleneck restricting prime editing activity.bioRxiv [Preprint]. 2023 Oct 29:2023.10.21.563443. doi: 10.1101/2023.10.21.563443. bioRxiv. 2023. Update in: Nat Biotechnol. 2025 Apr;43(4):539-544. doi: 10.1038/s41587-024-02405-x. PMID: 37904991 Free PMC article. Updated. Preprint.
References
Methods-only references:
MeSH terms
Substances
Grants and funding
- UH3 TR002668/TR/NCATS NIH HHS/United States
- R01 HL170629/HL/NHLBI NIH HHS/United States
- U54AI170856/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01 CA275945/CA/NCI NIH HHS/United States
- UH3TR002668/U.S. Department of Health & Human Services | NIH | National Center for Advancing Translational Sciences (NCATS)
- R01HL150669/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- R21 OD030004/OD/NIH HHS/United States
- R21OD030004/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R37 AI147868/AI/NIAID NIH HHS/United States
- U54HD0060848/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- R37AI147868/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- U54 AI170856/AI/NIAID NIH HHS/United States
- R35 HL140017/HL/NHLBI NIH HHS/United States
- UG3 TR002668/TR/NCATS NIH HHS/United States
- R35HL140017/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- COE-2020-967-M-N/JDRF/United States
- R01 HL150669/HL/NHLBI NIH HHS/United States
- R01 NS111990/NS/NINDS NIH HHS/United States
- R01AI150478/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R01GM150273/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R01 AI150478/AI/NIAID NIH HHS/United States
- R01 GM150273/GM/NIGMS NIH HHS/United States
- U54 HD060848/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

