Comparison of methods for building polygenic scores for diverse populations
- PMID: 39323095
- PMCID: PMC11532986
- DOI: 10.1016/j.xhgg.2024.100355
Comparison of methods for building polygenic scores for diverse populations
Abstract
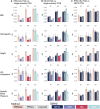
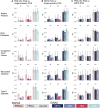
Polygenic scores (PGSs) are a promising tool for estimating individual-level genetic risk of disease based on the results of genome-wide association studies (GWASs). However, their promise has yet to be fully realized because most currently available PGSs were built with genetic data from predominantly European-ancestry populations, and PGS performance declines when scores are applied to target populations different from the populations from which they were derived. Thus, there is a great need to improve PGS performance in currently under-studied populations. In this work we leverage data from two large and diverse cohorts the Million Veterans Program (MVP) and All of Us (AoU), providing us the unique opportunity to compare methods for building PGSs for multi-ancestry populations across multiple traits. We build PGSs for five continuous traits and five binary traits using both multi-ancestry and single-ancestry approaches with popular Bayesian PGS methods and both MVP META GWAS results and population-specific GWAS results from the respective African, European, and Hispanic MVP populations. We evaluate these scores in three AoU populations genetically similar to the respective African, Admixed American, and European 1000 Genomes Project superpopulations. Using correlation-based tests, we make formal comparisons of the PGS performance across the multiple AoU populations. We conclude that approaches that combine GWAS data from multiple populations produce PGSs that perform better than approaches that utilize smaller single-population GWAS results matched to the target population, and specifically that multi-ancestry scores built with PRS-CSx outperform the other approaches in the three AoU populations.
Keywords: diverse populations; polygenic scores; underrepresented populations.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
-
- Khera A.V., Chaffin M., Aragam K.G., Haas M.E., Roselli C., Choi S.H., Natarajan P., Lander E.S., Lubitz S.A., Ellinor P.T., Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

