Proteomic exploration of potential blood biomarkers in haemophilic arthropathy
- PMID: 39323462
- PMCID: PMC11423339
- DOI: 10.1002/hsr2.70046
Proteomic exploration of potential blood biomarkers in haemophilic arthropathy
Abstract
Background and aims: The pathophysiology of haemophilic arthropathy (HA) is complex and largely undefined. Proteomic analyses provide insights into the intricate mechanisms of the HA.Our study aimed to identify differentially expressed proteins in relation to the severity of HA, explore their pathophysiological roles, and evaluate their potential as HA biomarkers.
Methods: Our cross-sectional observational study encompassed 30 HA patients and 15 healthy subjects. Plasma samples were pooled into three groups of 15 samples from those with severe haemophilic arthropathy (sHA), mild haemophilic arthropathy (mHA) and healthy controls. Proteomic analysis was performed using liquid chromatography-mass spectrometry. The severity of HA was assessed using the World Federation of Haemophilia Physical Examination Score and ultrasonography following the Haemophilia Early Arthropathy Detection with Ultrasound (HEAD-US) guidelines.
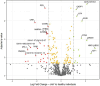
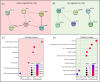
Results: A total of 788 proteins were identified, with 97% of the uniquely identified proteins being expressed in all analysed groups. We identified several up and downregulated proteins across the groups that were mainly related to inflammatory and immunity-modulating processes, as well as joint degeneration. We highlighted ten proteins relevant for the development of HA: cathepsin G, endoplasmic reticulum aminopeptidase 2, S100-A9, insulin-like growth factor I, apolipoprotein (a), osteopontin, pregnancy zone protein, cartilage oligomeric matrix protein, CD44, and cadherin-related family member 2.
Conclusion: Our analysis identified several proteins that shed further light on the distinctive pathogenesis of HA and could serve for biomarker research. However, these results need to be validated on a larger patient group.
Keywords: arthropathy; biomarkers; haemophilia; pathophysiology; proteomics.
© 2024 The Author(s). Health Science Reports published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Biomarkers and immunological parameters in haemophilia and rheumatoid arthritis patients: a comparative multiplexing laboratory study.Haemophilia. 2021 Jan;27(1):e119-e126. doi: 10.1111/hae.14200. Epub 2020 Nov 18. Haemophilia. 2021. PMID: 33210410
-
Tissue-inhibitors of metalloproteinase-1 and vascular-endothelial growth-factor in severe haemophilia A children on low dose prophylactic recombinant factor VIII: Relation to subclinical arthropathy.Haemophilia. 2020 Jul;26(4):607-614. doi: 10.1111/hae.14041. Epub 2020 May 23. Haemophilia. 2020. PMID: 32445517
-
An exploratory comparison of single intra-articular injection of platelet-rich plasma vs hyaluronic acid in treatment of haemophilic arthropathy of the knee.Haemophilia. 2019 May;25(3):484-492. doi: 10.1111/hae.13711. Epub 2019 Mar 13. Haemophilia. 2019. PMID: 30866117 Clinical Trial.
-
Blood-Induced Arthropathy: A Major Disabling Complication of Haemophilia.J Clin Med. 2023 Dec 30;13(1):225. doi: 10.3390/jcm13010225. J Clin Med. 2023. PMID: 38202232 Free PMC article. Review.
-
Haemophilic arthropathy: A narrative review on the use of intra-articular drugs for arthritis.Haemophilia. 2019 Nov;25(6):919-927. doi: 10.1111/hae.13857. Epub 2019 Oct 22. Haemophilia. 2019. PMID: 31639263 Review.
Cited by
-
RE: Proteomic Exploration of Potential Blood Biomarkers in Haemophilic Arthropathy.Health Sci Rep. 2024 Nov 29;7(12):e70226. doi: 10.1002/hsr2.70226. eCollection 2024 Dec. Health Sci Rep. 2024. PMID: 39619085 Free PMC article. No abstract available.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

