Structural and genomic evolutionary dynamics of Omicron variant of SARS-CoV-2 circulating in Madhya Pradesh, India
- PMID: 39323472
- PMCID: PMC11422100
- DOI: 10.3389/fmed.2024.1416006
Structural and genomic evolutionary dynamics of Omicron variant of SARS-CoV-2 circulating in Madhya Pradesh, India
Abstract
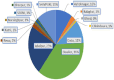
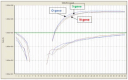
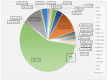
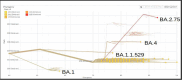
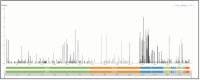
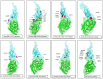
The SARS-CoV-2 Omicron (B.1.1.529) variant emerged in early November 2021 and its rapid spread created fear worldwide. This was attributed to its increased infectivity and escaping immune mechanisms. The spike protein of Omicron has more mutations (>30) than any other previous variants and was declared as the variant of concern (VOC) by the WHO. The concern among the scientific community was huge about this variant, and a piece of updated information on circulating viral strains is important in order to better understand the epidemiology, virus pathogenicity, transmission, therapeutic interventions, and vaccine development. A total of 710 samples were processed for sequencing and identification up to a resolution of sub-lineage. The sequence analysis revealed Omicron variant with distribution as follows: B.1.1, B.1.1.529, BA.1, BA.2, BA.2.10, BA.2.10.1, BA.2.23, BA.2.37, BA.2.38, BA.2.43, BA.2.74, BA.2.75, BA.2.76, and BA.4 sub-lineages. There is a shift noted in circulating lineage from BA.1 to BA.2 to BA.4 over a period from January to September 2022. Multiple signature mutations were identified in S protein T376A, D405N, and R408S mutations, which were new and common to all BA.2 variants. Additionally, R346T was seen in emerging BA.2.74 and BA.2.76 variants. The emerging BA.4 retained the common T376A, D405N, and R408S mutations of BA.2 along with a new mutation F486V. The samples sequenced were from different districts of Madhya Pradesh and showed a predominance of BA.2 and its variants circulating in this region. The current study identified circulation of BA.1 and BA.1.1 variants during initial phase. The predominant Delta strain of the second wave has been replaced by the Omicron variant in this region over a period of time. This study successfully deciphers the dynamics of the emergence and replacement of various sub-lineages of SARS-CoV-2 in central India on real real-time basis.
Keywords: BA.2; COVID-19; Epidemiology; Genome; Lineage; NGS.
Copyright © 2024 Dhankher, Yadav, Sharma, Gupta, Yadav, Dash and Parida.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Omicron BA.2 lineage predominance in severe acute respiratory syndrome coronavirus 2 positive cases during the third wave in North India.Front Med (Lausanne). 2022 Nov 2;9:955930. doi: 10.3389/fmed.2022.955930. eCollection 2022. Front Med (Lausanne). 2022. PMID: 36405589 Free PMC article.
-
Mutational characterization of Omicron SARS-CoV-2 lineages circulating in Chhattisgarh, a central state of India.Front Med (Lausanne). 2023 Jan 23;9:1082846. doi: 10.3389/fmed.2022.1082846. eCollection 2022. Front Med (Lausanne). 2023. PMID: 36755883 Free PMC article.
-
Mutational Analysis of Circulating Omicron SARS-CoV-2 Lineages in the Al-Baha Region of Saudi Arabia.J Multidiscip Healthc. 2023 Jul 27;16:2117-2136. doi: 10.2147/JMDH.S419859. eCollection 2023. J Multidiscip Healthc. 2023. PMID: 37529147 Free PMC article.
-
Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: Implications for immune escape and transmission.Rev Med Virol. 2022 Sep;32(5):e2381. doi: 10.1002/rmv.2381. Epub 2022 Jul 20. Rev Med Virol. 2022. PMID: 35856385 Free PMC article. Review.
-
The emergence of SARS-CoV-2 Omicron subvariants: current situation and future trends.Infez Med. 2022 Dec 1;30(4):480-494. doi: 10.53854/liim-3004-2. eCollection 2022. Infez Med. 2022. PMID: 36482957 Free PMC article. Review.
References
-
- World Health Organization (WHO) . WHO coronavirus (COVID-19) dashboard. Available at: https://covid19.who.int/ (Accessed February 20, 2024).
LinkOut - more resources
Full Text Sources
Miscellaneous

