Deciphering the Genetic Complexity of Classical Hodgkin Lymphoma: Insights and Effective Strategies
- PMID: 39323623
- PMCID: PMC11420564
- DOI: 10.2174/0113892029301904240513045755
Deciphering the Genetic Complexity of Classical Hodgkin Lymphoma: Insights and Effective Strategies
Abstract
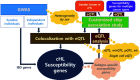
Understanding the genetics of susceptibility to classical Hodgkin lymphoma (cHL) is considerably limited compared to other cancers due to the rare Hodgkin and Reed-Sternberg (HRS) tumor cells, which coexist with the predominant non-malignant microenvironment. This article offers insights into genetic abnormalities in cHL, as well as nucleotide variants and their associated target genes, elucidated through recent technological advancements. Oncogenomes in HRS cells highlight the survival and proliferation of these cells through hyperactive signaling in specific pathways (e.g., NF-kB) and their interplay with microenvironmental cells (e.g., CD4+ T cells). In contrast, the susceptibility genes identified from genome-wide association studies and expression quantitative trait locus analyses only vaguely implicate their potential roles in susceptibility to more general cancers. To pave the way for the era of precision oncology, more intensive efforts are imperative, employing the following strategies: exploring genetic heterogeneity by gender and cHL subtype, investigating colocalization with various types of expression quantitative trait loci, and leveraging single-cell analysis. These approaches provide valuable perspectives for unraveling the genetic complexities of cHL.
Keywords: Hodgkin lymphoma; NF-kB; expression quantitative trait locus; genetic etiology; genetic heterogeneity; genetic lesion; genome-wide association study; hodgkin and reed-sternberg cell.
© 2024 The Author(s). Published by Bentham Science Publishers.
Conflict of interest statement
Chaeyoung Lee is the Editorial Advisory Board member of the journal Current Genomics.
Figures

Similar articles
-
Detection of genomic imbalances in microdissected Hodgkin and Reed-Sternberg cells of classical Hodgkin's lymphoma by array-based comparative genomic hybridization.Haematologica. 2008 Sep;93(9):1318-26. doi: 10.3324/haematol.12875. Epub 2008 Jul 18. Haematologica. 2008. PMID: 18641027
-
The impact of EBV and HIV infection on the microenvironmental niche underlying Hodgkin lymphoma pathogenesis.Int J Cancer. 2017 Mar 15;140(6):1233-1245. doi: 10.1002/ijc.30473. Epub 2016 Oct 27. Int J Cancer. 2017. PMID: 27750386 Review.
-
The biology of classical Hodgkin lymphoma.Semin Hematol. 2024 Aug;61(4):212-220. doi: 10.1053/j.seminhematol.2024.05.001. Epub 2024 May 8. Semin Hematol. 2024. PMID: 38824068 Review.
-
Molecular biology of Hodgkin lymphoma.Leukemia. 2021 Apr;35(4):968-981. doi: 10.1038/s41375-021-01204-6. Epub 2021 Mar 8. Leukemia. 2021. PMID: 33686198 Free PMC article. Review.
-
Molecular Evolution of Classic Hodgkin Lymphoma Revealed Through Whole-Genome Sequencing of Hodgkin and Reed Sternberg Cells.Blood Cancer Discov. 2023 May 1;4(3):208-227. doi: 10.1158/2643-3230.BCD-22-0128. Blood Cancer Discov. 2023. PMID: 36723991 Free PMC article.
References
-
- Küppers R., Rajewsky K., Zhao M., Simons G., Laumann R., Fischer R., Hansmann M.L. Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc. Natl. Acad. Sci. 1994;91(23):10962–10966. doi: 10.1073/pnas.91.23.10962. - DOI - PMC - PubMed
-
- Marafioti T., Hummel M., Foss H.D., Laumen H., Korbjuhn P., Anagnostopoulos I., Lammert H., Demel G., Theil J., Wirth T., Stein H. Hodgkin and Reed-Sternberg cells represent an expansion of a single clone originating from a germinal center B-cell with functional immunoglobulin gene rearrangements but defective immunoglobulin transcription. Blood. 2000;95(4):1443–1450. doi: 10.1182/blood.V95.4.1443.004k55_1443_1450. - DOI - PubMed
-
- Mack T.M., Cozen W., Shibata D.K., Weiss L.M., Nathwani B.N., Hernandez A.M., Taylor C.R., Hamilton A.S., Deapen D.M., Rappaport E.B. Concordance for Hodgkin’s disease in identical twins suggesting genetic susceptibility to the young-adult form of the disease. N. Engl. J. Med. 1995;332(7):413–419. doi: 10.1056/NEJM199502163320701. - DOI - PubMed
-
- Kharazmi E., Fallah M., Pukkala E., Olsen J.H., Tryggvadottir L., Sundquist K., Tretli S., Hemminki K. Risk of familial classical Hodgkin lymphoma by relationship, histology, age, and sex: A joint study from five Nordic countries. Blood. 2015;126(17):1990–1995. doi: 10.1182/blood-2015-04-639781. - DOI - PubMed
-
- Rudant J., Menegaux F., Leverger G., Baruchel A., Nelken B., Bertrand Y., Hartmann O., Pacquement H., Vérité C., Robert A., Michel G., Margueritte G., Gandemer V., Hémon D., Clavel J. Family history of cancer in children with acute leukemia, Hodgkin’s lymphoma or non-Hodgkin’s lymphoma: The ESCALE study (SFCE). Int. J. Cancer. 2007;121(1):119–126. doi: 10.1002/ijc.22624. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
