Genomic analysis of Enterococcus faecium from non-clinical settings: antimicrobial resistance, virulence, and clonal population in livestock and the urban environment
- PMID: 39323892
- PMCID: PMC11422121
- DOI: 10.3389/fmicb.2024.1466990
Genomic analysis of Enterococcus faecium from non-clinical settings: antimicrobial resistance, virulence, and clonal population in livestock and the urban environment
Abstract
Introduction: Enterococci are commensals of the gastrointestinal tract of humans and animals that evolved into opportunistic pathogens with high antimicrobial resistance and virulence. Multidrug-resistant Enterococcus is a major cause of hospital-acquired infections worldwide. For this reason, the characterization of non-clinical reservoirs of Enterococci and their epidemiological link to resistant hospital isolates is crucial for controlling their spread.
Methods: A total of 295 samples collected from livestock (pigs and cows, n = 135) and environment (public buses, passengers hands, and urban environments, n = 160) were screened for Enterococcus spp. E. faecium antimicrobial resistance profiles, virulence potential, and clonal population were further characterized.
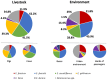
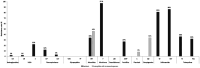
Results: Enterococci were detected in 90.5% (n = 267) of the samples, with a higher prevalence in livestock (100%) than the environment (82.5%, p < 0.0001), but none of the isolates exhibited vancomycin resistance. E. faecalis was the most prevalent species (51.7%), predominantly found in livestock (62.2%), while E. faecium was more common in the environment. Of the 59 E. faecium isolates, 78% showed resistance to ≥3 antibiotic classes and contained associated resistance genes, namely tetracyclines (tetM and tetL), beta-lactams (mutations in pbp5), and high-level resistance to aminoglycosides (ant(6)-Ia and aac(6')-aph(2″)). A wide array of virulence factors was detected among E. faecium, associated with adherence, biofilm formation, and adaptation to host response, while hospital-associated virulence markers, such as IS16, were less frequent, probably due to the non-clinical nature of the isolates. Clonal population analysis revealed a diverse E. faecium population. Although no direct epidemiological link could be traced between our isolates and specific clinical isolates, infection-associated genetic backgrounds were identified in non-clinical isolates: one isolate from pigs belonged to CC17 (ST32), while four isolates belonged to CC94, including one recovered from pigs (ST296), one from cows (ST2206), one from the urban environment (ST1205), and other from buses (ST800).
Discussion: This study underscores a high prevalence of clinically relevant Enterococcus species among healthy livestock and the environment. Despite the absence of vancomycin resistance and limited hospital infection-associated clonal lineages, the presence of E. faecium with significant virulence potential and resistance to critical antibiotics in human and veterinary medicine highlights the need for continuing surveillance of non-clinical reservoirs.
Keywords: Enterococcus faecium; Enterococcus spp.; aminoglycosides high-level resistance; ampicillin resistant E. faecium; antimicrobial resistance; environment; livestock; non-clinical enterococcus reservoirs.
Copyright © 2024 Lopes, de Lencastre and Conceição.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Abdullahi I. N., Lozano C., Juárez-Fernández G., Höfle U., Simón C., Rueda S., et al. (2023). Nasotracheal enterococcal carriage and resistomes: detection of optrA-, poxtA- and cfrD-carrying strains in migratory birds, livestock, pets, and in-contact humans in Spain. Eur. J. Clin. Microbiol. Infect. Dis. 42, 569–581. doi: 10.1007/s10096-023-04579-9, PMID: - DOI - PMC - PubMed
-
- Bender J. K., Cattoir V., Hegstad K., Sadowy E., Coque T. M., Westh H., et al. (2018a). Update on prevalence and mechanisms of resistance to linezolid, tigecycline and daptomycin in enterococci in Europe: towards a common nomenclature. Drug Resist. Up 40, 25–39. doi: 10.1016/j.drup.2018.10.002, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

