Alterations in the plasma proteome persist ten months after recovery from mild to moderate SARS-CoV-2 infection
- PMID: 39324144
- PMCID: PMC11422241
- DOI: 10.3389/fimmu.2024.1448780
Alterations in the plasma proteome persist ten months after recovery from mild to moderate SARS-CoV-2 infection
Abstract
Background: Limited data are available describing the effects of SARS-CoV-2 breakthrough infections on the plasma proteome.
Methods: PCR-positive SARS-CoV-2 patients, enrolled in a natural history study, underwent analysis of the plasma proteome. A prospective cohort of 66 unvaccinated and 24 vaccinated persons with different degrees of infection severity were evaluated acutely (within 40 days of symptom onset), and at three and ten months. Comparisons based on vaccination status alone and unsupervised hierarchical clustering were performed. A second cohort of vaccinated Omicron patients were evaluated acutely and at ten months.
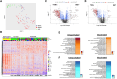
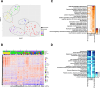
Results: Acutely, unvaccinated patients manifested overexpression of proteins involved in immune and inflammatory responses, while vaccinated patients exhibited adaptive immune responses without significant inflammation. At three and ten months, only unvaccinated patients had diminished but sustained inflammatory (C3b, CCL15, IL17RE) and immune responses (DEFA5,TREM1). Both groups had underexpression of pathways essential for cellular function, signaling, and angiogenesis (AKT1, MAPK14, HSPB1) across phases. Unsupervised clustering, based on protein expression, identified four groups of patients with variable vaccination rates demonstrating that additional clinical factors influence the plasma proteome. The proteome of vaccinated Omicron patients did not differ from vaccinated pre-Omicron patients.
Conclusions: Vaccination attenuates the inflammatory response to SARS-CoV-2 infection across phases. However, at ten months after symptom onset, changes in the plasma proteome persist in both vaccinated and unvaccinated individuals, which may be relevant to post-acute sequelae of SARS-CoV-2 and other viral infections associated with post-acute infection syndromes.
Keywords: SARS-CoV-2; breakthrough infections; inflammation; post-acute sequelae; proteomics; vaccination.
Copyright © 2024 Huapaya, Gairhe, Kanth, Tian, Demirkale, Regenold, Sun, Lynch, Luo, Forsberg, Dewar, Rehman, Li, Krack, Kuruppu, Aboye, Barnett, Strich, Davey, Childs, Chertow, Kovacs, Torabi-Parizi and Suffredini.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Bollyky TJ, Castro E, Aravkin AY, Bhangdia K, Dalos J, Hulland EN, et al. . Assessing COVID-19 pandemic policies and behaviours and their economic and educational trade-offs across US states from Jan 1, 2020, to July 31, 2022: an observational analysis. Lancet. (2023) 401(10385):1341–60. doi: 10.1016/S0140-6736(23)00461-0 - DOI - PMC - PubMed
-
- Filbin MR, Mehta A, Schneider AM, Kays KR, Guess JR, Gentili M, et al. . Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell-cell interactions. Cell Rep Med. (2021) 2:100287. doi: 10.1016/j.xcrm.2021.100287 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

