Heat shock factor ZmHsf17 positively regulates phosphatidic acid phosphohydrolase ZmPAH1 and enhances maize thermotolerance
- PMID: 39324623
- PMCID: PMC11714762
- DOI: 10.1093/jxb/erae406
Heat shock factor ZmHsf17 positively regulates phosphatidic acid phosphohydrolase ZmPAH1 and enhances maize thermotolerance
Abstract
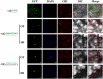
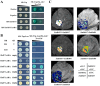
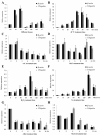
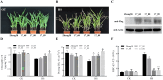
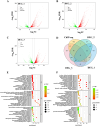
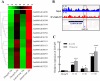
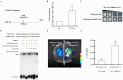
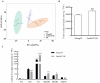
Heat stress adversely impacts plant growth, development, and grain yield. Heat shock factors (Hsf), especially the HsfA2 subclass, play a pivotal role in the transcriptional regulation of genes in response to heat stress. In this study, the coding sequence of maize ZmHsf17 was cloned. ZmHsf17 contained conserved domains including a DNA binding domain, oligomerization domain, and transcriptional activation domain. The protein was nuclear localized and had transcription activation activity. Yeast two-hybrid and split luciferase complementation assays confirmed the interaction of ZmHsf17 with members of the maize HsfA2 subclass. Overexpression of ZmHsf17 in maize significantly increased chlorophyll content and net photosynthetic rate, and enhanced the stability of cellular membranes. Through integrative analysis of ChIP-seq and RNA-seq datasets, ZmPAH1, encoding phosphatidic acid phosphohydrolase of lipid metabolic pathways, was identified as a target gene of ZmHsf17. The promoter fragment of ZmPAH1 was bound by ZmHsf17 in protein-DNA interaction experiments in vivo and in vitro. Lipidomic data also indicated that the overexpression of ZmHsf17 increased levels of some critical membrane lipid components of maize leaves under heat stress. This research provides new insights into the role of the ZmHsf17-ZmPAH1 module in regulating thermotolerance in maize.
Keywords: Heat shock factor; ZmPAH1; maize; membrane lipid component; thermotolerance; transcriptional regulation.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Conflict of interest statement
We declare that all authors have no conflict of interest.
Figures


Similar articles
-
Maize HSFA2 and HSBP2 antagonistically modulate raffinose biosynthesis and heat tolerance in Arabidopsis.Plant J. 2019 Oct;100(1):128-142. doi: 10.1111/tpj.14434. Epub 2019 Jul 12. Plant J. 2019. PMID: 31180156
-
ZmHsf05, a new heat shock transcription factor from Zea mays L. improves thermotolerance in Arabidopsis thaliana and rescues thermotolerance defects of the athsfa2 mutant.Plant Sci. 2019 Jun;283:375-384. doi: 10.1016/j.plantsci.2019.03.002. Epub 2019 Mar 18. Plant Sci. 2019. PMID: 31128708
-
Genome-wide identification, transcriptome analysis and alternative splicing events of Hsf family genes in maize.Sci Rep. 2020 May 15;10(1):8073. doi: 10.1038/s41598-020-65068-z. Sci Rep. 2020. PMID: 32415117 Free PMC article.
-
Heat Stress Responses and Thermotolerance in Maize.Int J Mol Sci. 2021 Jan 19;22(2):948. doi: 10.3390/ijms22020948. Int J Mol Sci. 2021. PMID: 33477941 Free PMC article. Review.
-
Impact of High-Temperature Stress on Maize Seed Setting: Cellular and Molecular Insights of Thermotolerance.Int J Mol Sci. 2025 Feb 2;26(3):1283. doi: 10.3390/ijms26031283. Int J Mol Sci. 2025. PMID: 39941051 Free PMC article. Review.
Cited by
-
Maize leaves salt-responsive genes revealed by comparative transcriptome of salt-tolerant and salt-sensitive cultivars during the seedling stage.PeerJ. 2025 Apr 10;13:e19268. doi: 10.7717/peerj.19268. eCollection 2025. PeerJ. 2025. PMID: 40226543 Free PMC article.
-
The Structural Deciphering of the α3 Helix Within ZmHsfA2'S DNA-Binding Domain for the Recognition of Heat Shock Elements in Maize.Plants (Basel). 2025 Jun 25;14(13):1950. doi: 10.3390/plants14131950. Plants (Basel). 2025. PMID: 40647959 Free PMC article.
-
Molecular Mechanisms of Plant Abiotic Stress Tolerance.Int J Mol Sci. 2025 Mar 18;26(6):2731. doi: 10.3390/ijms26062731. Int J Mol Sci. 2025. PMID: 40141369 Free PMC article.
References
-
- Andrási N, Pettkó-Szandtner A, Szabados L.. 2021. Diversity of plant heat shock factors: regulation, interactions, and functions. Journal of Experimental Botany 72, 1558–1575. - PubMed
-
- Balogh G, Péter M, Glatz A, Gombos I, Török Z, Horváth I, Harwood JL, Vígh L.. 2013. Key role of lipids in heat stress management. FEBS Letters 587, 1970–1980. - PubMed
-
- Balogi Z, Multhoff G, Jensen TK, Lloyd-Evans E, Yamashima T, Jäättelä M, Harwood JL, Vígh L.. 2019. Hsp70 interactions with membrane lipids regulate cellular functions in health and disease. Progress in Lipid Research 74, 18–30. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

