Widespread Adaptive Introgression of Major Histocompatibility Complex Genes across Vertebrate Hybrid Zones
- PMID: 39324637
- PMCID: PMC11472244
- DOI: 10.1093/molbev/msae201
Widespread Adaptive Introgression of Major Histocompatibility Complex Genes across Vertebrate Hybrid Zones
Abstract
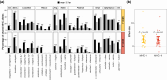
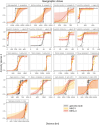
Interspecific introgression is a potentially important source of novel variation of adaptive significance. Although multiple cases of adaptive introgression are well documented, broader generalizations about its targets and mechanisms are lacking. Multiallelic balancing selection, particularly when acting through rare allele advantage, is an evolutionary mechanism expected to favor adaptive introgression. This is because introgressed alleles are likely to confer an immediate selective advantage, facilitating their establishment in the recipient species even in the face of strong genomic barriers to introgression. Vertebrate major histocompatibility complex genes are well-established targets of long-term multiallelic balancing selection, so widespread adaptive major histocompatibility complex introgression is expected. Here, we evaluate this hypothesis using data from 29 hybrid zones formed by fish, amphibians, squamates, turtles, birds, and mammals at advanced stages of speciation. The key prediction of more extensive major histocompatibility complex introgression compared to genome-wide introgression was tested with three complementary statistical approaches. We found evidence for widespread adaptive introgression of major histocompatibility complex genes, providing a link between the process of adaptive introgression and an underlying mechanism. Our work identifies major histocompatibility complex introgression as a general mechanism by which species can acquire novel, and possibly regain previously lost, variation that may enhance defense against pathogens and increase adaptive potential.
Keywords: MHC; adaptation; host–pathogen coevolution; hybridization; introgression.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of Interest The authors declare no conflicts of interest.
Figures




Similar articles
-
Massive introgression of major histocompatibility complex (MHC) genes in newt hybrid zones.Mol Ecol. 2019 Nov;28(21):4798-4810. doi: 10.1111/mec.15254. Epub 2019 Oct 21. Mol Ecol. 2019. PMID: 31574568
-
Interspecific introgression of MHC genes in Triturus newts: Evidence from multiple contact zones.Mol Ecol. 2023 Feb;32(4):867-880. doi: 10.1111/mec.16804. Epub 2022 Dec 16. Mol Ecol. 2023. PMID: 36458894 Free PMC article.
-
Widespread introgression of MHC genes in Iberian Podarcis lizards.Mol Ecol. 2023 Jul;32(14):4003-4017. doi: 10.1111/mec.16974. Epub 2023 May 4. Mol Ecol. 2023. PMID: 37143304
-
Evolution of MHC IIB Diversity Across Cichlid Fish Radiations.Genome Biol Evol. 2023 Jun 1;15(6):evad110. doi: 10.1093/gbe/evad110. Genome Biol Evol. 2023. PMID: 37314153 Free PMC article. Review.
-
Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation.Mol Ecol. 2013 Sep;22(18):4606-18. doi: 10.1111/mec.12415. Epub 2013 Aug 1. Mol Ecol. 2013. PMID: 23906376 Review.
Cited by
-
Human-mediated introgression and Varroa destructor shaped the genetic structure of honey bee populations in the Azores.Sci Rep. 2025 Jul 12;15(1):25217. doi: 10.1038/s41598-025-08950-y. Sci Rep. 2025. PMID: 40652044 Free PMC article.
References
-
- Aitken SN, Whitlock MC. Assisted gene flow to facilitate local adaptation to climate change. Annu Rev Ecol Evol Syst. 2013:44(1):367–388. 10.1146/annurev-ecolsys-110512-135747. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

