ATF6 Promotes Colorectal Cancer Growth and Stemness by Regulating the Wnt Pathway
- PMID: 39324706
- PMCID: PMC11492184
- DOI: 10.1158/2767-9764.CRC-24-0268
ATF6 Promotes Colorectal Cancer Growth and Stemness by Regulating the Wnt Pathway
Abstract
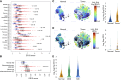
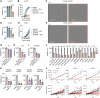
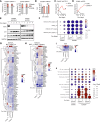
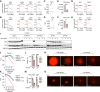
ATF6 intervention reduces colorectal cancer cell and organoid viability by interrupting dysregulated Wnt signaling, identifying a novel facilitator and potential therapeutic target in colorectal cancer.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
J.J. Rodvold reported being a full-time employee of Genentech Inc. at the time of this work. S.A. Marsters reports being a full-time employee of Genentech. I. Oikonomidi reports personal fees from Genentech Inc. during the conduct of the study. Z.D. Modrusan reports other support from Roche outside the submitted work. T.D. Wu reports other support from Genentech, Inc. during the conduct of the study and other support from Genentech, Inc. outside the submitted work. F. de Sousa e Melo reports owning shares of Roche and Amgen. A. Ashkenazi reports being a full-time employee of and other support from Genentech, Inc. outside the submitted work. No disclosures were reported by the other authors.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

