NOTUM-mediated WNT silencing drives extravillous trophoblast cell lineage development
- PMID: 39325428
- PMCID: PMC11459147
- DOI: 10.1073/pnas.2403003121
NOTUM-mediated WNT silencing drives extravillous trophoblast cell lineage development
Abstract
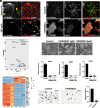
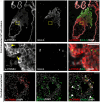
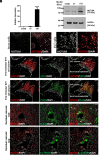
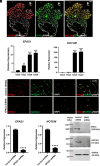
Trophoblast stem (TS) cells have the unique capacity to differentiate into specialized cell types, including extravillous trophoblast (EVT) cells. EVT cells invade into and transform the uterus where they act to remodel the vasculature facilitating the redirection of maternal nutrients to the developing fetus. Disruptions in EVT cell development and function are at the core of pregnancy-related disease. WNT-activated signal transduction is a conserved regulator of morphogenesis of many organ systems, including the placenta. In human TS cells, activation of canonical WNT signaling is critical for maintenance of the TS cell stem state and its downregulation accompanies EVT cell differentiation. We show that aberrant WNT signaling undermines EVT cell differentiation. Notum, palmitoleoyl-protein carboxylesterase (NOTUM), a negative regulator of canonical WNT signaling, was prominently expressed in first-trimester EVT cells developing in situ and up-regulated in EVT cells derived from human TS cells. Furthermore, NOTUM was required for optimal human TS cell differentiation to EVT cells. Activation of NOTUM in EVT cells is driven, at least in part, by endothelial Per-Arnt-Sim (PAS) domain 1 (also called hypoxia-inducible factor 2 alpha). Collectively, our findings indicate that canonical Wingless-related integration site (WNT) signaling is essential for maintenance of human trophoblast cell stemness and regulation of human TS cell differentiation. Downregulation of canonical WNT signaling via the actions of NOTUM is required for optimal EVT cell differentiation.
Keywords: NOTUM; WNT; extravillous trophoblast cells; placenta.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

Update of
-
NOTUM-MEDIATED WNT SILENCING DRIVES EXTRAVILLOUS TROPHOBLAST CELL LINEAGE DEVELOPMENT.bioRxiv [Preprint]. 2024 Aug 22:2024.02.13.579974. doi: 10.1101/2024.02.13.579974. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2024 Oct;121(40):e2403003121. doi: 10.1073/pnas.2403003121. PMID: 38405745 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- R01 HD112559/HD/NICHD NIH HHS/United States
- R01 HD020676/HD/NICHD NIH HHS/United States
- F32 HD096809/HD/NICHD NIH HHS/United States
- R01 HD099638/HD/NICHD NIH HHS/United States
- R00 HD107262/HD/NICHD NIH HHS/United States
- HD105734/HHS | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- K99 HD115834/HD/NICHD NIH HHS/United States
- KUMC Biomedical Research Training Program/KU | University of Kansas Medical Center (KUMC)
- P20 GM103418/GM/NIGMS NIH HHS/United States
- HD079363/HHS | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- HD020676/HHS | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- HD099638/HHS | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- HD096809/HHS | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- P01 HD079363/HD/NICHD NIH HHS/United States
- R01 HD105734/HD/NICHD NIH HHS/United States
- K99 HD107262/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources

