Knowledge Graphs for drug repurposing: a review of databases and methods
- PMID: 39325460
- PMCID: PMC11426166
- DOI: 10.1093/bib/bbae461
Knowledge Graphs for drug repurposing: a review of databases and methods
Abstract
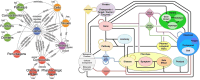
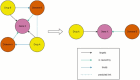
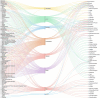
Drug repurposing has emerged as a effective and efficient strategy to identify new treatments for a variety of diseases. One of the most effective approaches for discovering potential new drug candidates involves the utilization of Knowledge Graphs (KGs). This review comprehensively explores some of the most prominent KGs, detailing their structure, data sources, and how they facilitate the repurposing of drugs. In addition to KGs, this paper delves into various artificial intelligence techniques that enhance the process of drug repurposing. These methods not only accelerate the identification of viable drug candidates but also improve the precision of predictions by leveraging complex datasets and advanced algorithms. Furthermore, the importance of explainability in drug repurposing is emphasized. Explainability methods are crucial as they provide insights into the reasoning behind AI-generated predictions, thereby increasing the trustworthiness and transparency of the repurposing process. We will discuss several techniques that can be employed to validate these predictions, ensuring that they are both reliable and understandable.
Keywords: artificial intelligence; drug repurposing; explainability; graph networks; knowledge graphs.
© The Author(s) 2024. Published by Oxford University Press.
Figures
Similar articles
-
Explainable drug repurposing via path based knowledge graph completion.Sci Rep. 2024 Jul 18;14(1):16587. doi: 10.1038/s41598-024-67163-x. Sci Rep. 2024. PMID: 39025897 Free PMC article.
-
An Analytical Review of Computational Drug Repurposing.IEEE/ACM Trans Comput Biol Bioinform. 2021 Mar-Apr;18(2):472-488. doi: 10.1109/TCBB.2019.2933825. Epub 2021 Apr 6. IEEE/ACM Trans Comput Biol Bioinform. 2021. PMID: 31403439 Review.
-
Drug repurposing using artificial intelligence, molecular docking, and hybrid approaches: A comprehensive review in general diseases vs Alzheimer's disease.Comput Methods Programs Biomed. 2025 Apr;261:108604. doi: 10.1016/j.cmpb.2025.108604. Epub 2025 Jan 13. Comput Methods Programs Biomed. 2025. PMID: 39826482 Review.
-
Applications of Artificial Intelligence in Drug Repurposing.Adv Sci (Weinh). 2025 Apr;12(14):e2411325. doi: 10.1002/advs.202411325. Epub 2025 Mar 6. Adv Sci (Weinh). 2025. PMID: 40047357 Free PMC article. Review.
-
HGTDR: Advancing drug repurposing with heterogeneous graph transformers.Bioinformatics. 2024 Jul 1;40(7):btae349. doi: 10.1093/bioinformatics/btae349. Bioinformatics. 2024. PMID: 38913860 Free PMC article.
Cited by
-
circ2LO: Identification of CircRNA Based on the LucaOne Large Model.Genes (Basel). 2025 Mar 31;16(4):413. doi: 10.3390/genes16040413. Genes (Basel). 2025. PMID: 40282373 Free PMC article.
-
AI-enabled drug prediction and gene network analysis reveal therapeutic use of vorinostat for Rett Syndrome in preclinical models.Commun Med (Lond). 2025 Jul 1;5(1):249. doi: 10.1038/s43856-025-00975-8. Commun Med (Lond). 2025. PMID: 40595330 Free PMC article.
-
Drug repurposing for Alzheimer's disease using a graph-of-thoughts based large language model to infer drug-disease relationships in a comprehensive knowledge graph.BioData Min. 2025 Aug 5;18(1):51. doi: 10.1186/s13040-025-00466-5. BioData Min. 2025. PMID: 40764997 Free PMC article.
-
From Lab to Clinic: How Artificial Intelligence (AI) Is Reshaping Drug Discovery Timelines and Industry Outcomes.Pharmaceuticals (Basel). 2025 Jun 30;18(7):981. doi: 10.3390/ph18070981. Pharmaceuticals (Basel). 2025. PMID: 40732273 Free PMC article. Review.
-
Adaptive multi-view learning method for enhanced drug repurposing using chemical-induced transcriptional profiles, knowledge graphs, and large language models.J Pharm Anal. 2025 Jun;15(6):101275. doi: 10.1016/j.jpha.2025.101275. Epub 2025 Mar 21. J Pharm Anal. 2025. PMID: 40678475 Free PMC article.
References
-
- Wouters OJ, McKee M, Luyten J. Estimated Research and Development investment needed to bring a new medicine to market, 2009-2018. JAMA 2020; 323:844. Available from: /pmc/articles/PMC7054832/ /pmc/articles/PMC7054832/?report=abstract https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7054832/. 10.1001/jama.2020.1166. - DOI - PMC - PubMed
-
- Sun D, Gao W, Hu H. et al. .. Why 90. Acta Pharma Sin B 2022; 7:3049. Available from: /pmc/articles/PMC9293739/ /pmc/articles/PMC9293739/?report=abstract https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9293739/. - PMC - PubMed
-
- Drug repurposing and repositioning: Workshop summary. Drug Repurpos Reposition 2014; 8:1. Available from: https://pubmed.ncbi.nlm.nih.gov/24872991/. - PubMed
-
- Lexchin J, Mintzes B. Semaglutide: a new drug for the treatment of obesity. Drug Ther Bull 2023; 61:182–8. Available from: https://dtb.bmj.com/content/61/12/182.abstract. 10.1136/dtb.2023.000007. - DOI - PubMed
-
- Jarada TN, Rokne JG, Alhajj R. A review of computational drug repositioning: strategies, approaches, opportunities, challenges, and directions. J Chem 2020; 12:12. Available from: https://api.semanticscholar.org/CorpusID:220680962. 10.1186/s13321-020-00450-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

