Analysis of CNS autoimmunity in genetically diverse mice reveals unique phenotypes and mechanisms
- PMID: 39325545
- PMCID: PMC11601571
- DOI: 10.1172/jci.insight.184138
Analysis of CNS autoimmunity in genetically diverse mice reveals unique phenotypes and mechanisms
Abstract
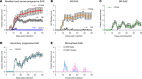
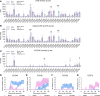
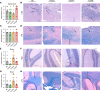
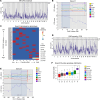
Multiple sclerosis (MS) is a complex disease with significant heterogeneity in disease course and progression. Genetic studies have identified numerous loci associated with MS risk, but the genetic basis of disease progression remains elusive. To address this, we leveraged the Collaborative Cross (CC), a genetically diverse mouse strain panel, and experimental autoimmune encephalomyelitis (EAE). The 32 CC strains studied captured a wide spectrum of EAE severity, trajectory, and presentation, including severe-progressive, monophasic, relapsing remitting, and axial rotary-EAE (AR-EAE), accompanied by distinct immunopathology. Sex differences in EAE severity were observed in 6 strains. Quantitative trait locus analysis revealed distinct genetic linkage patterns for different EAE phenotypes, including EAE severity and incidence of AR-EAE. Machine learning-based approaches prioritized candidate genes for loci underlying EAE severity (Abcc4 and Gpc6) and AR-EAE (Yap1 and Dync2h1). This work expands the EAE phenotypic repertoire and identifies potentially novel loci controlling unique EAE phenotypes, supporting the hypothesis that heterogeneity in MS disease course is driven by genetic variation.
Keywords: Autoimmunity; Genetic variation; Genetics; Mouse models; Multiple sclerosis.
Conflict of interest statement
Figures




Update of
-
Probing the basis of disease heterogeneity in multiple sclerosis using genetically diverse mice.bioRxiv [Preprint]. 2024 Jun 4:2024.06.03.597205. doi: 10.1101/2024.06.03.597205. bioRxiv. 2024. Update in: JCI Insight. 2024 Nov 8;9(21):e184138. doi: 10.1172/jci.insight.184138. PMID: 38895248 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

