A novel viral RNA detection method based on the combined use of trans-acting ribozymes and HCR-FRET analyses
- PMID: 39325749
- PMCID: PMC11426510
- DOI: 10.1371/journal.pone.0310171
A novel viral RNA detection method based on the combined use of trans-acting ribozymes and HCR-FRET analyses
Abstract
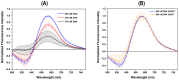
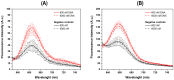
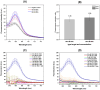
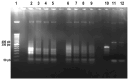
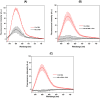
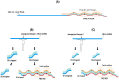
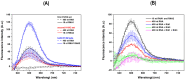
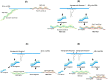
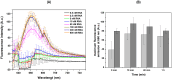
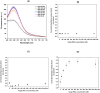
The diagnoses of retroviruses are essential for controlling the rapid spread of pandemics. However, the real-time Reverse Transcriptase quantitative Polymerase Chain Reaction (RT-qPCR), which has been the gold standard for identifying viruses such as SARS-CoV-2 in the early stages of infection, is associated with high costs and logistical challenges. To innovate in viral RNA detection a novel molecular approach for detecting SARS-CoV-2 viral RNA, as a proof of concept, was developed. This method combines specific viral gene analysis, trans-acting ribozymes, and Fluorescence Resonance Energy Transfer (FRET)-based hybridization of fluorescent DNA hairpins. In this molecular mechanism, SARS-CoV-2 RNA is specifically recognized and cleaved by ribozymes, releasing an initiator fragment that triggers a hybridization chain reaction (HCR) with DNA hairpins containing fluorophores, leading to a FRET process. A consensus SARS-CoV-2 RNA target sequence was identified, and specific ribozymes were designed and transcribed in vitro to cleave the viral RNA into fragments. DNA hairpins labeled with Cy3/Cy5 fluorophores were then designed and synthesized for HCR-FRET assays targeting the RNA fragment sequences resulting from ribozyme cleavage. The results demonstrated that two of the three designed ribozymes effectively cleaved the target RNA within 10 minutes. Additionally, DNA hairpins labeled with Cy3/Cy5 pairs efficiently detected target RNA specifically and triggered detectable HCR-FRET reactions. This method is versatile and can be adapted for use with other viruses. Furthermore, the design and construction of a DIY photo-fluorometer prototype enabled us to explore the development of a simple and cost-effective point-of-care detection method based on digital image analysis.
Copyright: © 2024 Ferreira da Silva et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
A CRISPR/Cas13a-based and hybridization chain reaction coupled evanescent wave biosensor for SARS-CoV-2 gene detection.Analyst. 2025 Mar 24;150(7):1367-1376. doi: 10.1039/d4an01584c. Analyst. 2025. PMID: 40035496
-
Target-independent hybridization chain reaction-fluorescence resonance energy transfer for sensitive assay of ctDNA based on Cas12a.Anal Chim Acta. 2023 Jun 22;1261:341170. doi: 10.1016/j.aca.2023.341170. Epub 2023 Apr 4. Anal Chim Acta. 2023. PMID: 37147050
-
Immuno-HCR based on contact quenching and fluorescence resonance energy transfer for sensitive and low background detection of Escherichia coli O157:H7.Food Chem. 2021 Jan 1;334:127568. doi: 10.1016/j.foodchem.2020.127568. Epub 2020 Jul 17. Food Chem. 2021. PMID: 32712489
-
Advancements in detection of SARS-CoV-2 infection for confronting COVID-19 pandemics.Lab Invest. 2022 Jan;102(1):4-13. doi: 10.1038/s41374-021-00663-w. Epub 2021 Sep 8. Lab Invest. 2022. PMID: 34497366 Free PMC article. Review.
-
Progress of the Detection Methods for SARS-CoV-2.Clin Lab. 2024 Aug 1;70(8). doi: 10.7754/Clin.Lab.2024.231148. Clin Lab. 2024. PMID: 39193975 Review.
References
-
- Prevention. CfDCa. Antigen Testing Guidelines. 2022 [https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-gu....
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

