DNA methylation profiling identifies TBKBP1 as potent amplifier of cytotoxic activity in CMV-specific human CD8+ T cells
- PMID: 39325839
- PMCID: PMC11460711
- DOI: 10.1371/journal.ppat.1012581
DNA methylation profiling identifies TBKBP1 as potent amplifier of cytotoxic activity in CMV-specific human CD8+ T cells
Abstract
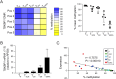
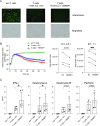
Epigenetic mechanisms stabilize gene expression patterns during CD8+ T cell differentiation. Although adoptive transfer of virus-specific T cells is clinically applied to reduce the risk of virus infection or reactivation in immunocompromised individuals, the DNA methylation pattern of virus-specific CD8+ T cells is largely unknown. Hence, we here performed whole-genome bisulfite sequencing of cytomegalovirus-specific human CD8+ T cells and found that they display a unique DNA methylation pattern consisting of 79 differentially methylated regions (DMRs) when compared to memory CD8+ T cells. Among the top demethylated DMRs in cytomegalovirus-specific CD8+ T cells was TBKBP1, coding for TBK-binding protein 1 that can interact with TANK-binding kinase 1 (TBK1) and mediate pro-inflammatory responses in innate immune cells downstream of intracellular virus sensing. Since TBKBP1 has not yet been reported in T cells, we aimed to unravel its role in virus-specific CD8+ T cells. TBKBP1 demethylation in terminal effector CD8+ T cells correlated with higher TBKBP1 expression at both mRNA and protein level, independent of alternative splicing of TBKBP1 transcripts. Notably, the distinct DNA methylation patterns in CD8+ T cell subsets was stable upon long-term in vitro culture. TBKBP1 overexpression resulted in enhanced TBK1 phosphorylation upon stimulation of CD8+ T cells and significantly improved their virus neutralization capacity. Collectively, our data demonstrate that TBKBP1 modulates virus-specific CD8+ T cell responses and could be exploited as therapeutic target to improve adoptive T cell therapies.
Copyright: © 2024 Yu et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Therapeutic Vaccination of Hematopoietic Cell Transplantation Recipients Improves Protective CD8 T-Cell Immunotherapy of Cytomegalovirus Infection.Front Immunol. 2021 Aug 19;12:694588. doi: 10.3389/fimmu.2021.694588. eCollection 2021. Front Immunol. 2021. PMID: 34489940 Free PMC article.
-
[Monitoring of cytomegalovirus-specific CD4+ and CD8+ T cell responses by cytokine flow cytometry in renal transplant recipients].Mikrobiyol Bul. 2016 Apr;50(2):224-35. Mikrobiyol Bul. 2016. PMID: 27175495 Turkish.
-
Abundant cytomegalovirus (CMV) reactive clonotypes in the CD8(+) T cell receptor alpha repertoire following allogeneic transplantation.Clin Exp Immunol. 2016 Jun;184(3):389-402. doi: 10.1111/cei.12770. Epub 2016 Mar 8. Clin Exp Immunol. 2016. PMID: 26800118 Free PMC article.
-
Molecular characterization of HCMV-specific immune responses: Parallels between CD8(+) T cells, CD4(+) T cells, and NK cells.Eur J Immunol. 2015 Sep;45(9):2433-45. doi: 10.1002/eji.201545495. Epub 2015 Aug 24. Eur J Immunol. 2015. PMID: 26228786 Review.
-
Cytomegalovirus infection and progressive differentiation of effector-memory T cells.F1000Res. 2018 Sep 26;7:F1000 Faculty Rev-1554. doi: 10.12688/f1000research.15753.1. eCollection 2018. F1000Res. 2018. PMID: 30345004 Free PMC article. Review.
References
-
- Takenaka K, Nishida T, Asano-Mori Y, Oshima K, Ohashi K, Mori T, et al.. Cytomegalovirus reactivation after allogeneic hematopoietic stem cell transplantation is associated with a reduced risk of relapse in patients with acute myeloid leukemia who survived to day 100 after transplantation: the Japan Society for Hematopoietic Cell Transplantation Transplantation-related Complication working group. Biol Blood Marrow Transplant. 2015;21(11):2008–16. doi: 10.1016/j.bbmt.2015.07.019 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

