NAD+ Metabolism and Mitochondrial Activity in the Aged Oocyte: Focus on the Effects of NAMPT Stimulation
- PMID: 39325941
- PMCID: PMC11567263
- DOI: 10.14336/AD.2024.0241
NAD+ Metabolism and Mitochondrial Activity in the Aged Oocyte: Focus on the Effects of NAMPT Stimulation
Abstract
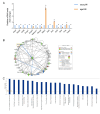
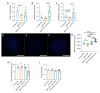
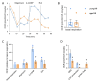
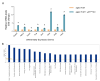
The ovary experiences an age-dependent decline starting during the fourth decade of life. Ovarian aging is the predominant factor driving female reproductive aging. Modern trend to postpone childbearing age contributes to reduced fertility and natality worldwide. Recently, the beneficial role of NAD+ precursors on the maintenance of oocyte competence and female fertility affected by aging has emerged. Nevertheless, age-related changes in NAD+ regulatory network have not been investigated so far. In this context, our goal was to investigate changes induced by the aging process in the expression level of genes participating in NAD+ biosynthetic and NAD+ consuming pathways and in the cellular bioenergetics in the mouse oocyte. From Ingenuity Pathway Analysis (IPA) it emerged that aging caused the downregulation of all cellular pathways for NAD+ synthesis (Kynurenine pathway, Preiss-Handler pathway and NAD+ salvage pathway) and deeply influenced the activity of NAD+-dependent enzymes, i.e. PARPs and SIRTs, with effects on many cellular functions including compromised ROS detoxification. Considering that NAMPT, the rate-limiting enzyme of NAD+ salvage pathway, was deregulated, aged oocytes were matured in the presence of P7C3, NAMPT activator. P7C3 improved spindle assembly and mitochondrial bioenergetics and reduced mitochondrial proton leak. Moreover, P7C3 influenced gene expression of NAD+ regulatory network, with Sirt1 as the central node of IPA-interfered target gene network. Finally, P7C3 effectively counteracted oocyte alterations induced by exposure to oxidative stress. Our study contributes to establish effective NAD+ boosting interventions to alleviate the effects of advanced maternal age on fertility and explore their potential in redox-related fertility disorders.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures









References
-
- Kulikova VA, Gromyko DV, Nikiforov AA (2018). The Regulatory Role of NAD in Human and Animal Cells. Biochem Mosc, 83:800-812. - PubMed
-
- Olgun A (2009). Converting NADH to NAD+ by nicotinamide nucleotide transhydrogenase as a novel strategy against mitochondrial pathologies during aging. Biogerontology, 10:531-534. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
