Insight into noncanonical small noncoding RNAs in Influenza A virus infection
- PMID: 39326700
- PMCID: PMC11466576
- DOI: 10.1016/j.virusres.2024.199474
Insight into noncanonical small noncoding RNAs in Influenza A virus infection
Abstract
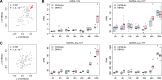
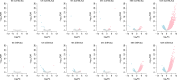
Influenza A virus (IAV) induces acute respiratory infections in birds and various mammals, including humans, and presents a significant global public health concern, with considerable economic consequences. Recently, researchers have shown keen interest in noncanonical small noncoding RNAs (sncRNAs) as carriers of epigenetic information, including tRNA-derived small RNAs (tsRNAs), rRNA-derived small RNA (rsRNAs), and Y RNA-derived small RNAs (ysRNAs). Particularly, tsRNAs and rsRNAs are detected in diverse species and demonstrate evolutionary conservation. We analyzed sncRNAs sequencing data in the pulmonary tissue of two genetically distinct mouse strains, C57BL/6J and DBA/2J, to explore strain-specific variations of sncRNAs in response to IAV infection. We systematically compiled information on noncanonical sncRNAs in these two strains and investigated the tsRNAs/rsRNAs/ysRNAs profiles influenced by IAV infection. Specifically, four noncanonical sncRNA families, including rsRNA-12S, GtsRNA-Arg-CCT, GtsRNA-Arg-TCT, and GtsRNA-Lys-TTT, exhibited upregulation upon IAV infection. Notably, DBA/2J mice showed earlier systemic differential expression of noncanonical sncRNAs after IAV infection compared to C57BL/6J mice. Additionally, our study revealed a strain-specific biogenesis of MtsRNAs in response to IAV infection. Also, distinct co-expression patterns of MtsRNAs were observed between C57BL/6J and DBA/2J mice, with DBA/2J mice showing broader positive co-expression of MtsRNAs with various sncRNA families compared to C57BL/6J mice. Our study provides a novel insight into noncanonical sncRNAs and their implications in IAV pathology and mouse strain specificity.
Keywords: Influenza A virus; Small noncoding RNAs; rsRNAs; tsRNAs; ysRNAs.
Copyright © 2024. Published by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Infection- and procedure-dependent effects on pulmonary gene expression in the early phase of influenza A virus infection in mice.BMC Microbiol. 2013 Dec 17;13:293. doi: 10.1186/1471-2180-13-293. BMC Microbiol. 2013. PMID: 24341411 Free PMC article.
-
PANDORA-Seq unveils the hidden small noncoding RNA landscape in atherosclerosis of LDL receptor-deficient mice.J Lipid Res. 2023 Apr;64(4):100352. doi: 10.1016/j.jlr.2023.100352. Epub 2023 Mar 4. J Lipid Res. 2023. PMID: 36871792 Free PMC article.
-
Plasma-Derived Exosomal SncRNA as a Promising Diagnostic Biomarker for Early Detection of HBV-Related Acute-on-Chronic Liver Failure.Front Cell Infect Microbiol. 2022 Jul 7;12:923300. doi: 10.3389/fcimb.2022.923300. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35873157 Free PMC article.
-
Exploring the expanding universe of small RNAs.Nat Cell Biol. 2022 Apr;24(4):415-423. doi: 10.1038/s41556-022-00880-5. Epub 2022 Apr 12. Nat Cell Biol. 2022. PMID: 35414016 Free PMC article. Review.
-
The significance of small noncoding RNAs in the pathogenesis of cardiovascular diseases.Genes Dis. 2024 May 30;12(4):101342. doi: 10.1016/j.gendis.2024.101342. eCollection 2025 Jul. Genes Dis. 2024. PMID: 40247912 Free PMC article. Review.
References
-
- Alberts R., Srivastava B., Wu H., Viegas N., Geffers R., Klawonn F., Novoselova N., do Valle T.Z., Panthier J.J., Schughart K. Gene expression changes in the host response between resistant and susceptible inbred mouse strains after Influenza A infection. Microbes Infect. 2010;12(4):309–318. - PubMed
-
- Casalino-Matsuda S.M., Chen F., Gonzalez-Gonzalez F.J., Matsuda H., Nair A., Abdala-Valencia H., Budinger G.R.S., Dong J.T., Beitel G.J., Sporn P.H. Myeloid Zfhx3 deficiency protects against hypercapnia-induced suppression of host defense against Influenza A virus. JCI Insight. 2024;9(4) - PMC - PubMed
-
- Chen Q., Li D., Jiang L., Wu Y., Yuan H., Shi G., Liu F., Wu P., Jiang K. Biological functions and clinical significance of tRNA-derived small fragment (tsRNA) in tumors: current state and future perspectives. Cancer Lett. 2024;587 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

