Unveiling the Virome of Wild Birds: Exploring CRESS-DNA Viral Dark Matter
- PMID: 39327897
- PMCID: PMC11463337
- DOI: 10.1093/gbe/evae206
Unveiling the Virome of Wild Birds: Exploring CRESS-DNA Viral Dark Matter
Abstract
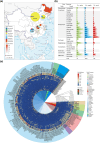
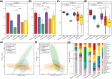
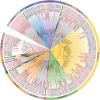
Amid global health concerns and the constant threat of zoonotic diseases, this study delves into the diversity of circular replicase-encoding single-stranded DNA (CRESS-DNA) viruses within Chinese wild bird populations. Employing viral metagenomics to tackle the challenge of "viral dark matter," the research collected and analyzed 3,404 cloacal swab specimens across 26 bird families. Metagenomic analysis uncovered a rich viral landscape, with 67.48% of reads classified as viral dark matter, spanning multiple taxonomic levels. Notably, certain viral families exhibited host-specific abundance patterns, with Galliformes displaying the highest diversity. Diversity analysis categorized samples into distinct groups, revealing significant differences in viral community structure, particularly noting higher diversity in terrestrial birds compared to songbirds and unique diversity in migratory birds versus perching birds. The identification of ten novel Circoviridae viruses, seven Smacoviridae viruses, and 167 Genomoviridae viruses, along with 100 unclassified CRESS-DNA viruses, underscores the expansion of knowledge on avian-associated circular DNA viruses. Phylogenetic and structural analyses of Rep proteins offered insights into evolutionary relationships and potential functional variations among CRESS-DNA viruses. In conclusion, this study significantly enhances our understanding of the avian virome, shedding light on the intricate relationships between viral communities and host characteristics in Chinese wild bird populations. The diverse array of CRESS-DNA viruses discovered opens avenues for future research into viral evolution, spread factors, and potential ecosystem impacts.
Keywords: Cressdnaviricota; dark matter; metagenomic; wild bird.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of Interest None declared.
Figures




Similar articles
-
Circular replication-associated protein encoding DNA viruses identified in the faecal matter of various animals in New Zealand.Infect Genet Evol. 2016 Sep;43:151-64. doi: 10.1016/j.meegid.2016.05.008. Epub 2016 May 20. Infect Genet Evol. 2016. PMID: 27211884
-
Pervasive Chimerism in the Replication-Associated Proteins of Uncultured Single-Stranded DNA Viruses.Viruses. 2018 Apr 10;10(4):187. doi: 10.3390/v10040187. Viruses. 2018. PMID: 29642587 Free PMC article.
-
Genetic Diversity and Characterization of Circular Replication (Rep)-Encoding Single-Stranded (CRESS) DNA Viruses.Microbiol Spectr. 2022 Dec 21;10(6):e0105722. doi: 10.1128/spectrum.01057-22. Epub 2022 Nov 8. Microbiol Spectr. 2022. PMID: 36346238 Free PMC article.
-
Eukaryotic Circular Rep-Encoding Single-Stranded DNA (CRESS DNA) Viruses: Ubiquitous Viruses With Small Genomes and a Diverse Host Range.Adv Virus Res. 2019;103:71-133. doi: 10.1016/bs.aivir.2018.10.001. Epub 2018 Dec 5. Adv Virus Res. 2019. PMID: 30635078 Review.
-
Towards an understanding of the avian virome.J Gen Virol. 2020 Aug;101(8):785-790. doi: 10.1099/jgv.0.001447. Epub 2020 Jun 9. J Gen Virol. 2020. PMID: 32519942 Free PMC article.
Cited by
-
Potential New Avian Species as Carriers of Diverse Circoviruses.Pathogens. 2025 May 28;14(6):540. doi: 10.3390/pathogens14060540. Pathogens. 2025. PMID: 40559547 Free PMC article.
-
Identification and characterization of novel CRESS-DNA viruses in the human respiratory tract.Virol J. 2025 Jun 30;22(1):211. doi: 10.1186/s12985-025-02742-6. Virol J. 2025. PMID: 40588733 Free PMC article.
References
-
- Briddon RW, Martin DP, Roumagnac P, Navas-Castillo J, Fiallo-Olivé E, Moriones E, Lett JM, Zerbini FM, Varsani A. Alphasatellitidae: a new family with two subfamilies for the classification of geminivirus- and nanovirus-associated alphasatellites. Arch Virol. 2018:163(9):2587–2600. 10.1007/s00705-018-3854-2. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

